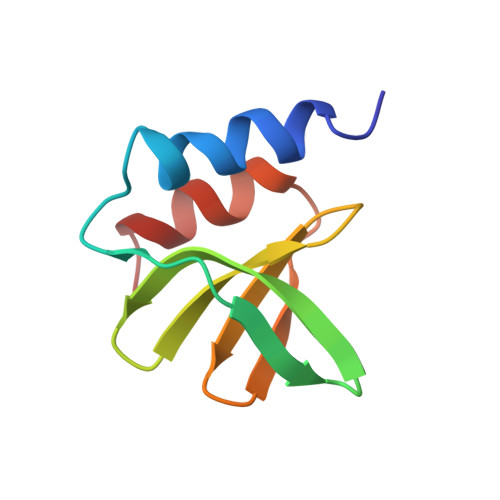
SARS-unique fold in the Rousettus bat coronavirus HKU9.
Hammond, R.G., Tan, X., Johnson, M.A.(2017) Protein Sci 26: 1726-1737
- PubMed: 28580734
- DOI: https://doi.org/10.1002/pro.3208
- Primary Citation of Related Structures:
5UTV - PubMed Abstract:
The coronavirus nonstructural protein 3 (nsp3) is a multifunctional protein that comprises multiple structural domains. This protein assists viral polyprotein cleavage, host immune interference, and may play other roles in genome replication or transcription. Here, we report the solution NMR structure of a protein from the "SARS-unique region" of the bat coronavirus HKU9. The protein contains a frataxin fold or double-wing motif, which is an α + β fold that is associated with protein/protein interactions, DNA binding, and metal ion binding. High structural similarity to the human severe acute respiratory syndrome (SARS) coronavirus nsp3 is present. A possible functional site that is conserved among some betacoronaviruses has been identified using bioinformatics and biochemical analyses. This structure provides strong experimental support for the recent proposal advanced by us and others that the "SARS-unique" region is not unique to the human SARS virus, but is conserved among several different phylogenetic groups of coronaviruses and provides essential functions.
- Department of Chemistry, University of Alabama at Birmingham, Birmingham, Alabama, 35294.
Organizational Affiliation: