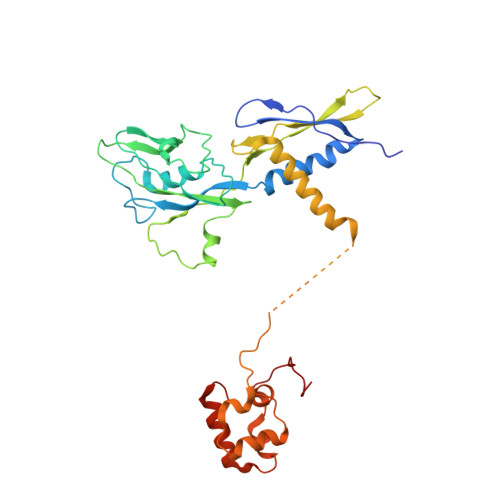
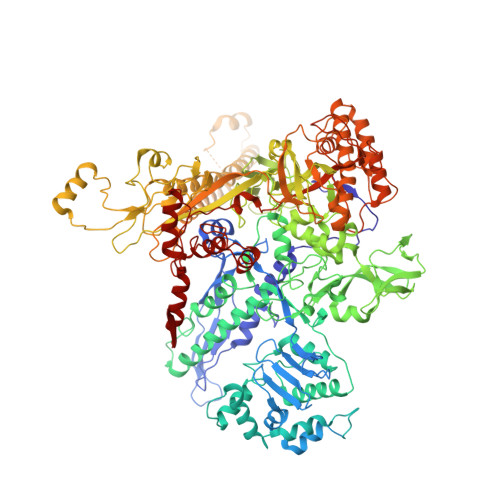
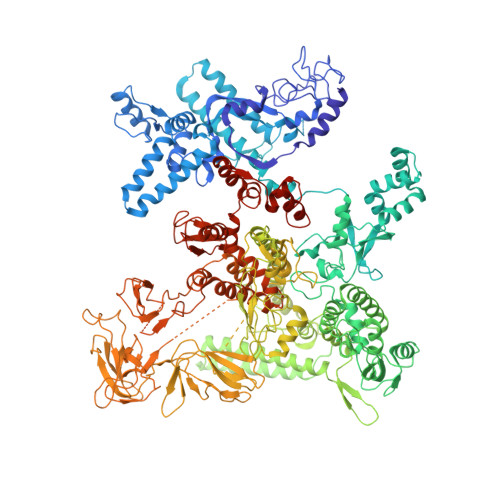
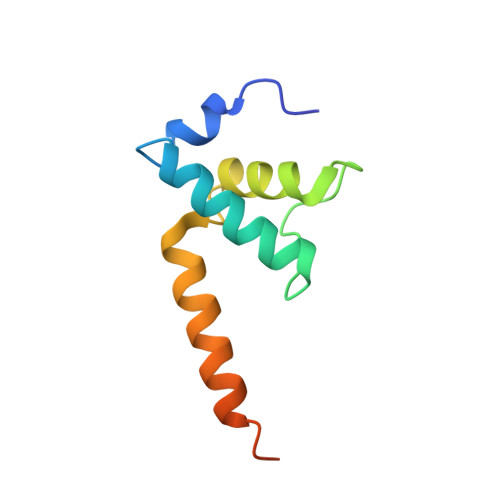
Crystal structure of Aquifex aeolicus sigma (N) bound to promoter DNA and the structure of sigma (N)-holoenzyme.
Campbell, E.A., Kamath, S., Rajashankar, K.R., Wu, M., Darst, S.A.(2017) Proc Natl Acad Sci U S A 114: E1805-E1814
- PubMed: 28223493
- DOI: https://doi.org/10.1073/pnas.1619464114
- Primary Citation of Related Structures:
5UI5, 5UI8 - PubMed Abstract:
The bacterial σ factors confer promoter specificity to the RNA polymerase (RNAP). One alternative σ factor, σ N , is unique in its structure and functional mechanism, forming transcriptionally inactive promoter complexes that require activation by specialized AAA + ATPases. We report a 3.4-Å resolution X-ray crystal structure of a σ N fragment in complex with its cognate promoter DNA, revealing the molecular details of promoter recognition by σ N The structure allowed us to build and refine an improved σ N -holoenzyme model based on previously published 3.8-Å resolution X-ray data. The improved σ N -holoenzyme model reveals a conserved interdomain interface within σ N that, when disrupted by mutations, leads to transcription activity without activator intervention (so-called bypass mutants). Thus, the structure and stability of this interdomain interface are crucial for the role of σ N in blocking transcription activity and in maintaining the activator sensitivity of σ N .
- Laboratory for Molecular Biophysics, The Rockefeller University, New York, NY 10065.
Organizational Affiliation: