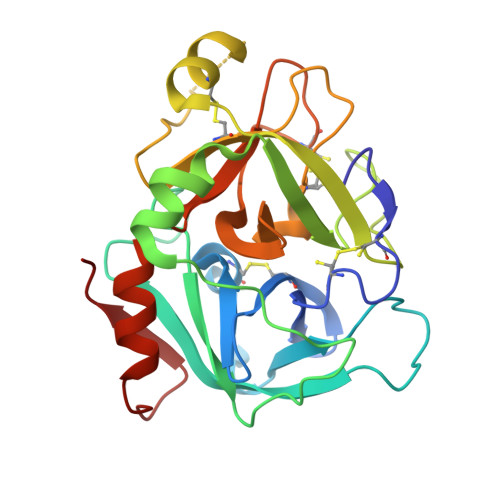
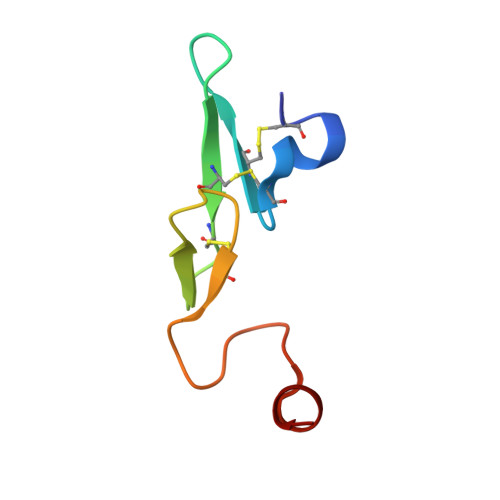
Neutral macrocyclic factor VIIa inhibitors.
Wurtz, N.R., Parkhurst, B.L., DeLucca, I., Glunz, P.W., Jiang, W., Zhang, X., Cheney, D.L., Bozarth, J.M., Rendina, A.R., Wei, A., Harper, T., Luettgen, J.M., Wu, Y., Wong, P.C., Seiffert, D.A., Wexler, R.R., Priestley, E.S.(2017) Bioorg Med Chem Lett 27: 2650-2654
- PubMed: 28460818
- DOI: https://doi.org/10.1016/j.bmcl.2017.04.008
- Primary Citation of Related Structures:
5U6J - PubMed Abstract:
Factor VIIa (FVIIa) inhibitors have shown strong antithrombotic efficacy in preclinical thrombosis models with limited bleeding liabilities. Discovery of potent, orally active FVIIa inhibitors has been largely unsuccessful due to the requirement of a basic P1 group to interact with Asp189 in the S1 binding pocket, limiting their membrane permeability. We have combined recently reported neutral P1 binding substituents with a highly optimized macrocyclic chemotype to produce FVIIa inhibitors with low nanomolar potency and enhanced permeability.
- Bristol-Myers Squibb Research and Development, Princeton, NJ 08534, United States. Electronic address: nicholas.wurtz@bms.com.
Organizational Affiliation: