Structure and function of the mycobacterial transcription initiation complex with the essential regulator RbpA.
Hubin, E.A., Fay, A., Xu, C., Bean, J.M., Saecker, R.M., Glickman, M.S., Darst, S.A., Campbell, E.A.(2017) Elife 6
- PubMed: 28067618
- DOI: https://doi.org/10.7554/eLife.22520
- Primary Citation of Related Structures:
5TW1 - PubMed Abstract:
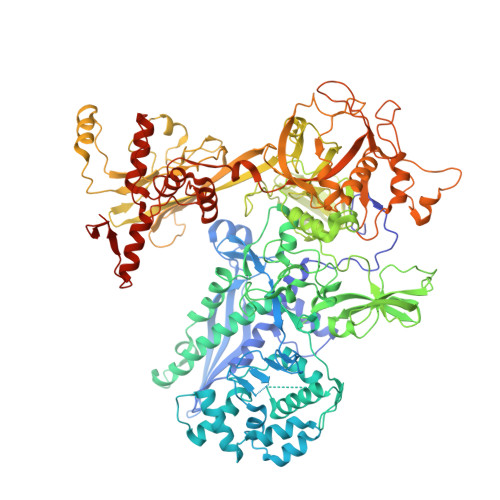
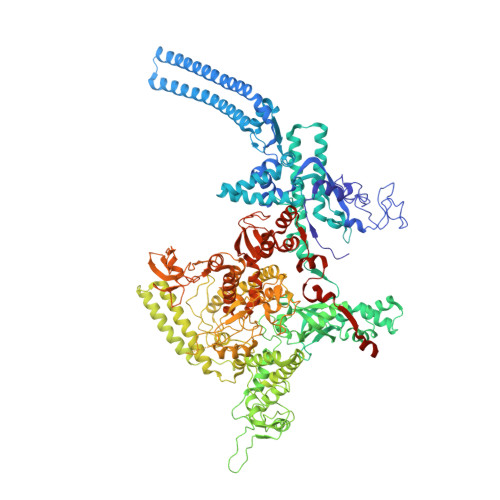
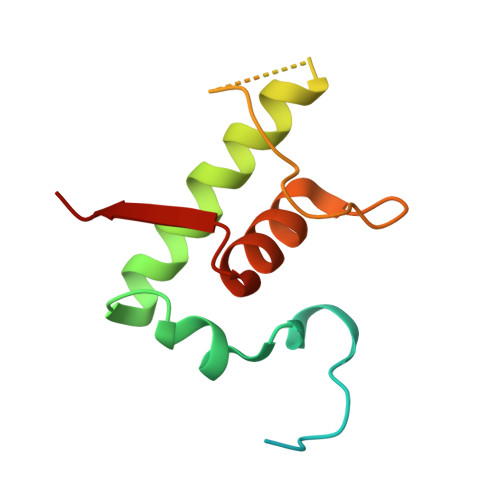
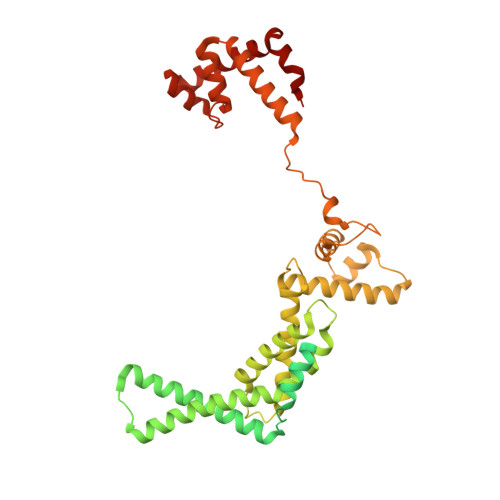
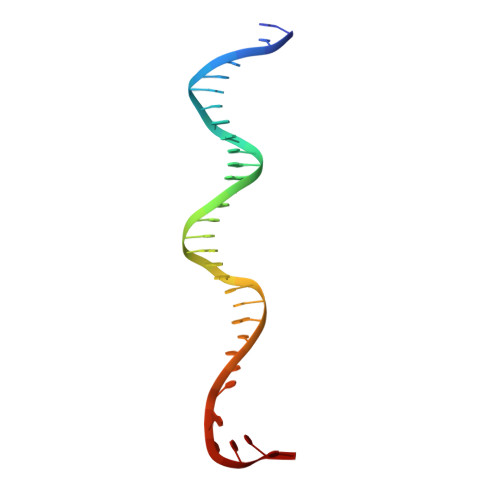
RbpA and CarD are essential transcription regulators in mycobacteria. Mechanistic analyses of promoter open complex (RPo) formation establish that RbpA and CarD cooperatively stimulate formation of an intermediate (RP2) leading to RPo; formation of RP2 is likely a bottleneck step at the majority of mycobacterial promoters. Once RPo forms, CarD also disfavors its isomerization back to RP2. We determined a 2.76 Å-resolution crystal structure of a mycobacterial transcription initiation complex (TIC) with RbpA as well as a CarD/RbpA/TIC model. Both CarD and RbpA bind near the upstream edge of the -10 element where they likely facilitate DNA bending and impede transcription bubble collapse. In vivo studies demonstrate the essential role of RbpA, show the effects of RbpA truncations on transcription and cell physiology, and indicate additional functions for RbpA not evident in vitro. This work provides a framework to understand the control of mycobacterial transcription by RbpA and CarD.
- The Rockefeller University, New York, United States.
Organizational Affiliation: