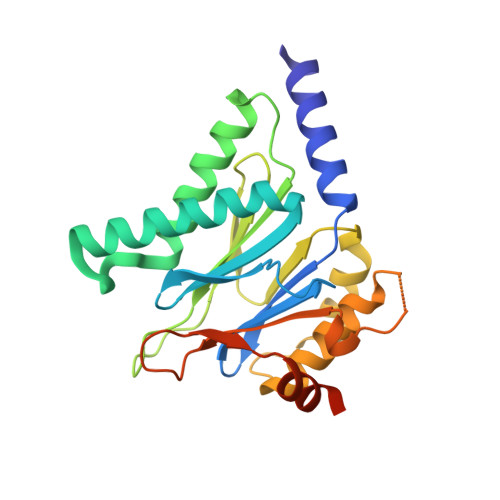
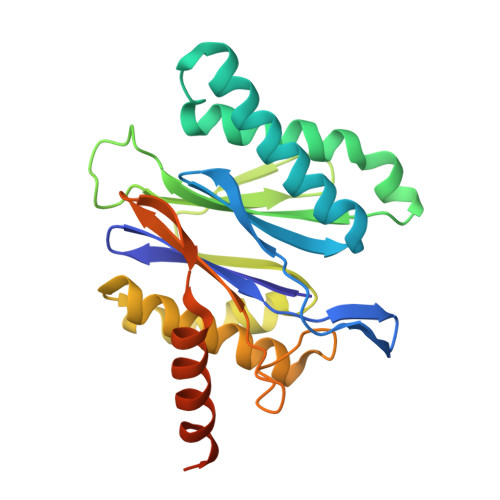
Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Hsu, H.C., Singh, P.K., Fan, H., Wang, R., Sukenick, G., Nathan, C., Lin, G., Li, H.(2017) Biochemistry 56: 324-333
- PubMed: 27976853
- DOI: https://doi.org/10.1021/acs.biochem.6b01107
- Primary Citation of Related Structures:
5THO, 5TRG, 5TRR, 5TRS, 5TRY, 5TS0 - PubMed Abstract:
The Mycobacterium tuberculosis (Mtb) 20S proteasome is vital for the pathogen to survive under nitrosative stress in vitro and to persist in mice. To qualify for drug development, inhibitors targeting Mtb 20S must spare both the human constitutive proteasome (c-20S) and immunoproteasome (i-20S). We recently reported members of a family of noncovalently binding dipeptide proteasome inhibitors that are highly potent and selective for Mtb 20S over human c-20S and i-20S. To understand the structural basis of their potency and selectivity, we have studied the structure-activity relationship of six derivatives and solved their cocrystal structures with Mtb 20S. The dipeptide inhibitors form an antiparallel β-strand with the active site β-strands. Selectivity is conferred by several features of Mtb 20S relative to its mouse counterparts, including a larger S1 pocket, additional hydrogen bonds in the S3 pocket, and hydrophobic interactions in the S4 pocket. Serine-20 and glutamine-22 of Mtb 20S interact with the dipeptides and confer Mtb-specific inhibition over c-20S and i-20S. The Mtb 20S and mammalian i-20S have a serine-27 that interacts strongly with the dipeptides, potentially explaining the higher inhibitory activity of the dipeptides toward i-20S over c-20S. This detailed structural knowledge will aid in optimizing the dipeptides as anti-tuberculosis drugs.
- Van Andel Research Institute , Grand Rapids, Michigan 49503, United States.
Organizational Affiliation: