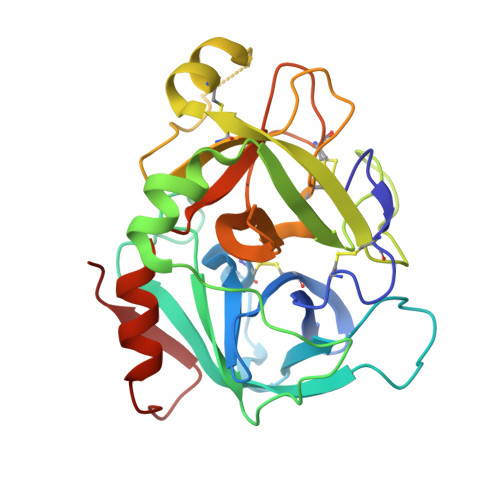
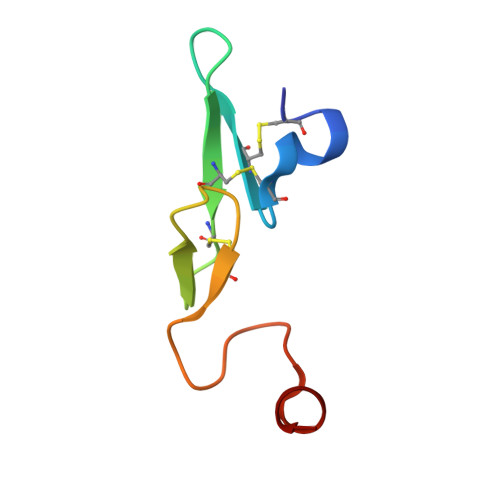
Design and Synthesis of Novel Meta-Linked Phenylglycine Macrocyclic FVIIa Inhibitors.
Richter, J.M., Cheney, D.L., Bates, J.A., Wei, A., Luettgen, J.M., Rendina, A.R., Harper, T.M., Narayanan, R., Wong, P.C., Seiffert, D., Wexler, R.R., Priestley, E.S.(2017) ACS Med Chem Lett 8: 67-72
- PubMed: 28105277
- DOI: https://doi.org/10.1021/acsmedchemlett.6b00375
- Primary Citation of Related Structures:
5TQE, 5TQF, 5TQG - PubMed Abstract:
Two novel series of meta-linked phenylglycine-based macrocyclic FVIIa inhibitors have been designed to improve the rodent metabolic stability and PK observed with the precursor para-linked phenylglycine macrocycles. Through iterative structure-based design and optimization, the TF/FVIIa K i was improved to subnanomolar levels with good clotting activity, metabolic stability, and permeability.
- Research & Development, Bristol-Myers Squibb , Hopewell, New Jersey 08534, United States.
Organizational Affiliation: