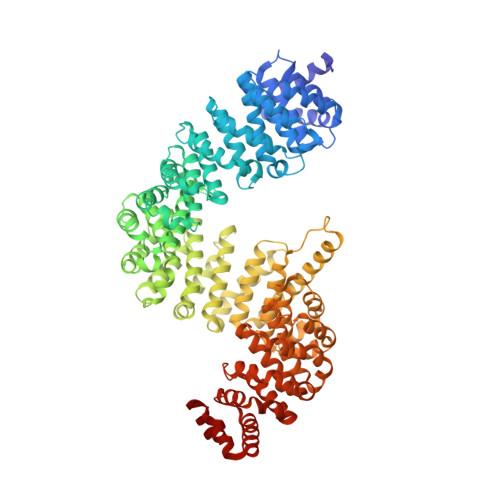
Molecular basis for protection of ribosomal protein L4 from cellular degradation.
Huber, F.M., Hoelz, A.(2017) Nat Commun 8: 14354-14354
- PubMed: 28148929
- DOI: https://doi.org/10.1038/ncomms14354
- Primary Citation of Related Structures:
5TQB, 5TQC - PubMed Abstract:
Eukaryotic ribosome biogenesis requires the nuclear import of ∼80 nascent ribosomal proteins and the elimination of excess amounts by the cellular degradation machinery. Assembly chaperones recognize nascent unassembled ribosomal proteins and transport them together with karyopherins to their nuclear destination. We report the crystal structure of ribosomal protein L4 (RpL4) bound to its dedicated assembly chaperone of L4 (Acl4), revealing extensive interactions sequestering 70 exposed residues of the extended RpL4 loop. The observed molecular recognition fundamentally differs from canonical promiscuous chaperone-substrate interactions. We demonstrate that the eukaryote-specific RpL4 extension harbours overlapping binding sites for Acl4 and the nuclear transport factor Kap104, facilitating its continuous protection from the cellular degradation machinery. Thus, Acl4 serves a dual function to facilitate nuclear import and simultaneously protect unassembled RpL4 from the cellular degradation machinery.
- Division of Chemistry and Chemical Engineering, California Institute of Technology, 1200 East California Boulevard, Pasadena, California 91125, USA.
Organizational Affiliation: