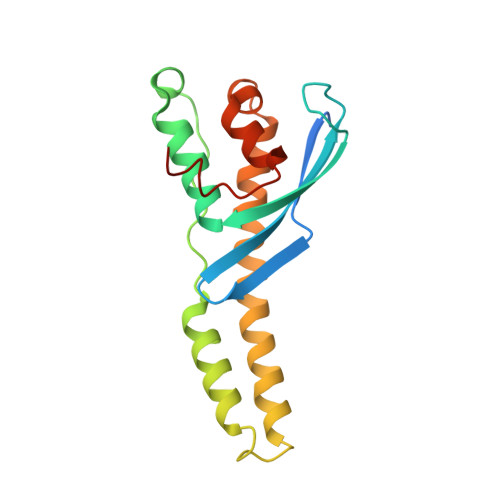
Chlamydia interfere with an interaction between the mannose-6-phosphate receptor and sorting nexins to counteract host restriction.
Elwell, C.A., Czudnochowski, N., von Dollen, J., Johnson, J.R., Nakagawa, R., Mirrashidi, K., Krogan, N.J., Engel, J.N., Rosenberg, O.S.(2017) Elife 6
- PubMed: 28252385
- DOI: https://doi.org/10.7554/eLife.22709
- Primary Citation of Related Structures:
5TP1 - PubMed Abstract:
Chlamydia trachomatis is an obligate intracellular pathogen that resides in a membrane-bound compartment, the inclusion. The bacteria secrete a unique class of proteins, Incs, which insert into the inclusion membrane and modulate the host-bacterium interface. We previously reported that IncE binds specifically to the Sorting Nexin 5 Phox domain (SNX5-PX) and disrupts retromer trafficking. Here, we present the crystal structure of the SNX5-PX:IncE complex, showing IncE bound to a unique and highly conserved hydrophobic groove on SNX5. Mutagenesis of the SNX5-PX:IncE binding surface disrupts a previously unsuspected interaction between SNX5 and the cation-independent mannose-6-phosphate receptor (CI-MPR). Addition of IncE peptide inhibits the interaction of CI-MPR with SNX5. Finally, C. trachomatis infection interferes with the SNX5:CI-MPR interaction, suggesting that IncE and CI-MPR are dependent on the same binding surface on SNX5. Our results provide new insights into retromer assembly and underscore the power of using pathogens to discover disease-related cell biology.
- Department of Medicine, University of California, San Francisco, San Francisco, United States.
Organizational Affiliation: