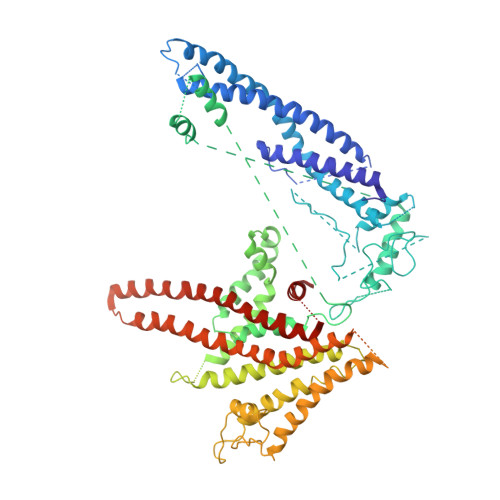
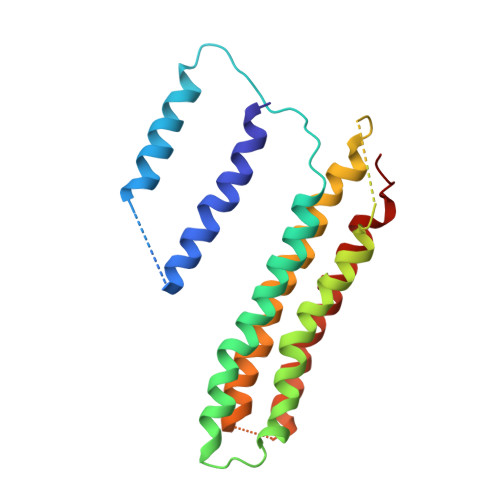
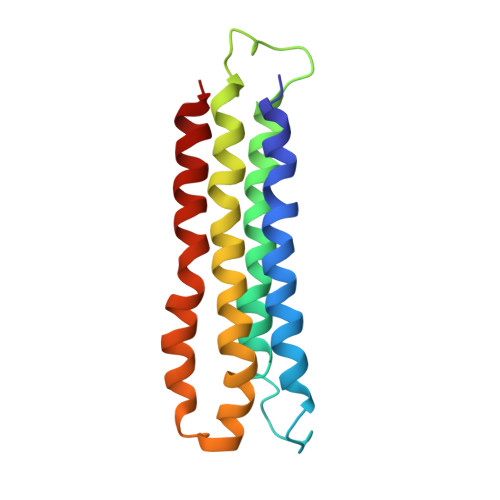
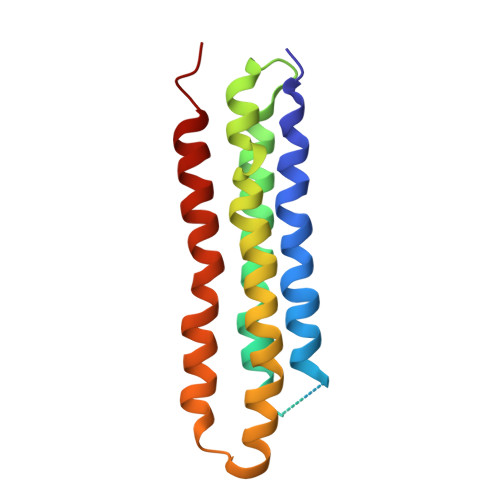
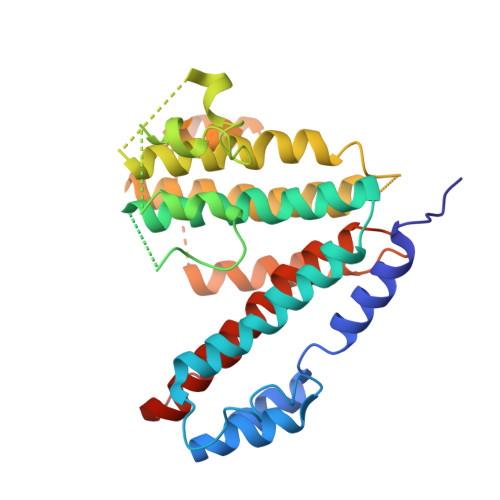
Atomic model for the membrane-embedded VO motor of a eukaryotic V-ATPase.
Mazhab-Jafari, M.T., Rohou, A., Schmidt, C., Bueler, S.A., Benlekbir, S., Robinson, C.V., Rubinstein, J.L.(2016) Nature 539: 118-122
- PubMed: 27776355
- DOI: https://doi.org/10.1038/nature19828
- Primary Citation of Related Structures:
5TJ5 - PubMed Abstract:
Vacuolar-type ATPases (V-ATPases) are ATP-powered proton pumps involved in processes such as endocytosis, lysosomal degradation, secondary transport, TOR signalling, and osteoclast and kidney function. ATP hydrolysis in the soluble catalytic V 1 region drives proton translocation through the membrane-embedded V O region via rotation of a rotor subcomplex. Variability in the structure of the intact enzyme has prevented construction of an atomic model for the membrane-embedded motor of any rotary ATPase. We induced dissociation and auto-inhibition of the V 1 and V O regions of the V-ATPase by starving the yeast Saccharomyces cerevisiae, allowing us to obtain a ~3.9-Å resolution electron cryomicroscopy map of the V O complex and build atomic models for the majority of its subunits. The analysis reveals the structures of subunits ac 8 c'c″de and a protein that we identify and propose to be a new subunit (subunit f). A large cavity between subunit a and the c-ring creates a cytoplasmic half-channel for protons. The c-ring has an asymmetric distribution of proton-carrying Glu residues, with the Glu residue of subunit c″ interacting with Arg735 of subunit a. The structure suggests sequential protonation and deprotonation of the c-ring, with ATP-hydrolysis-driven rotation causing protonation of a Glu residue at the cytoplasmic half-channel and subsequent deprotonation of a Glu residue at a luminal half-channel.
- Molecular Structure and Function Program, The Hospital for Sick Children, Toronto, Ontario M5G 0A4, Canada.
Organizational Affiliation: