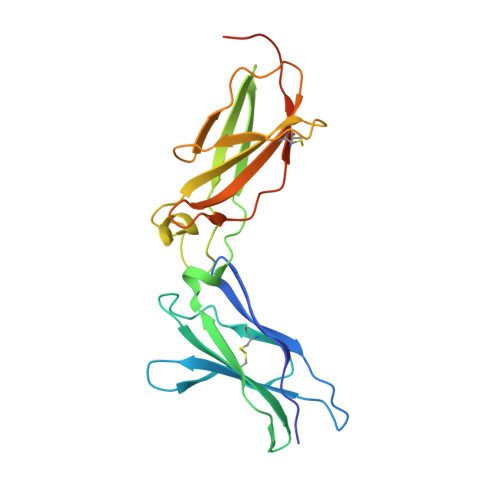
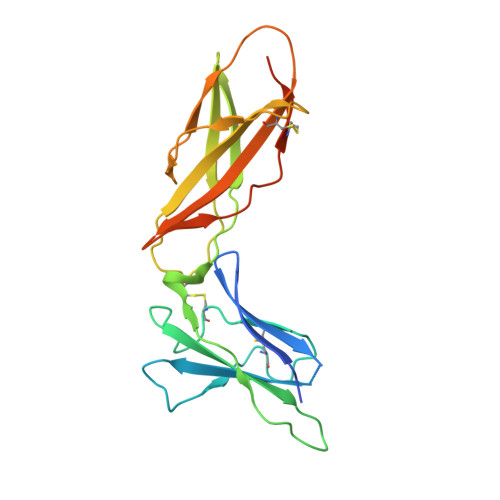
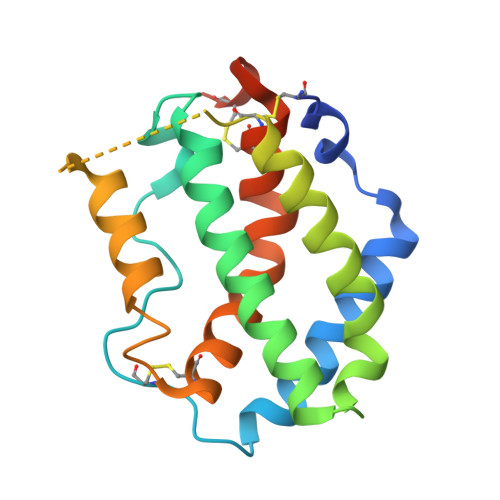
The IFN-lambda-IFN-lambda R1-IL-10R beta Complex Reveals Structural Features Underlying Type III IFN Functional Plasticity.
Mendoza, J.L., Schneider, W.M., Hoffmann, H.H., Vercauteren, K., Jude, K.M., Xiong, A., Moraga, I., Horton, T.M., Glenn, J.S., de Jong, Y.P., Rice, C.M., Garcia, K.C.(2017) Immunity 46: 379-392
- PubMed: 28329704
- DOI: https://doi.org/10.1016/j.immuni.2017.02.017
- Primary Citation of Related Structures:
5T5W - PubMed Abstract:
Type III interferons (IFN-λs) signal through a heterodimeric receptor complex composed of the IFN-λR1 subunit, specific for IFN-λs, and interleukin-10Rβ (IL-10Rβ), which is shared by multiple cytokines in the IL-10 superfamily. Low affinity of IL-10Rβ for cytokines has impeded efforts aimed at crystallizing cytokine-receptor complexes. We used yeast surface display to engineer a higher-affinity IFN-λ variant, H11, which enabled crystallization of the ternary complex. The structure revealed that IL-10Rβ uses a network of tyrosine residues as hydrophobic anchor points to engage IL-10 family cytokines that present complementary hydrophobic binding patches, explaining its role as both a cross-reactive but cytokine-specific receptor. H11 elicited increased anti-proliferative and antiviral activities in vitro and in vivo. In contrast, engineered higher-affinity type I IFNs did not increase antiviral potency over wild-type type I IFNs. Our findings provide insight into cytokine recognition by the IL-10R family and highlight the plasticity of type III interferon signaling and its therapeutic potential.
- Howard Hughes Medical Institute, Department of Molecular and Cellular Physiology and Department of Structural Biology, Stanford University School of Medicine, Stanford, CA 94305, USA.
Organizational Affiliation: