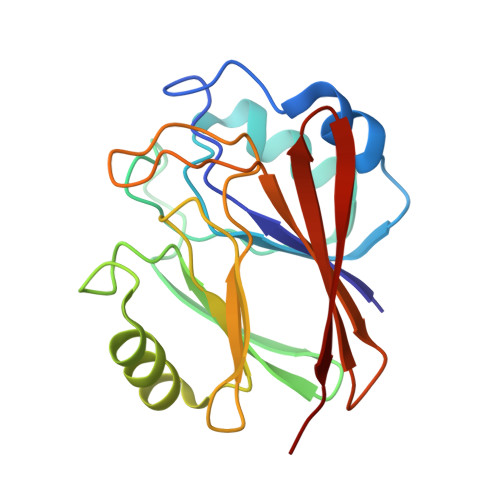
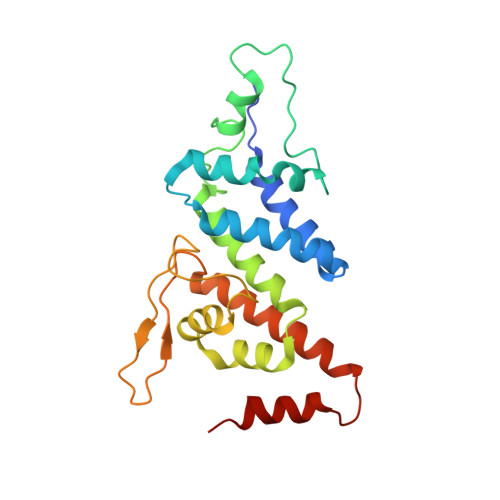
Molecular mechanism for the subversion of the retromer coat by the Legionella effector RidL.
Romano-Moreno, M., Rojas, A.L., Williamson, C.D., Gershlick, D.C., Lucas, M., Isupov, M.N., Bonifacino, J.S., Machner, M.P., Hierro, A.(2017) Proc Natl Acad Sci U S A 114: E11151-E11160
- PubMed: 29229824
- DOI: https://doi.org/10.1073/pnas.1715361115
- Primary Citation of Related Structures:
5OSH, 5OSI, 5OT4 - PubMed Abstract:
Microbial pathogens employ sophisticated virulence strategies to cause infections in humans. The intracellular pathogen Legionella pneumophila encodes RidL to hijack the host scaffold protein VPS29, a component of retromer and retriever complexes critical for endosomal cargo recycling. Here, we determined the crystal structure of L. pneumophila RidL in complex with the human VPS29-VPS35 retromer subcomplex. A hairpin loop protruding from RidL inserts into a conserved pocket on VPS29 that is also used by cellular ligands, such as Tre-2/Bub2/Cdc16 domain family member 5 (TBC1D5) and VPS9-ankyrin repeat protein for VPS29 binding. Consistent with the idea of molecular mimicry in protein interactions, RidL outcompeted TBC1D5 for binding to VPS29. Furthermore, the interaction of RidL with retromer did not interfere with retromer dimerization but was essential for association of RidL with retromer-coated vacuolar and tubular endosomes. Our work thus provides structural and mechanistic evidence into how RidL is targeted to endosomal membranes.
- Structural Biology Unit, Centro de Investigación Cooperativa en Biociencias, 48160 Derio, Spain.
Organizational Affiliation: