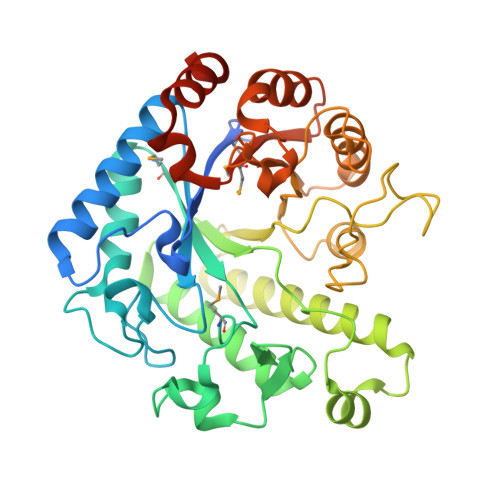
The molecular structure of the glycoside hydrolase domain of Cwp19 from Clostridium difficile.
Bradshaw, W.J., Kirby, J.M., Roberts, A.K., Shone, C.C., Acharya, K.R.(2017) FEBS J 284: 4343-4357
- PubMed: 29083543
- DOI: https://doi.org/10.1111/febs.14310
- Primary Citation of Related Structures:
5OQ2, 5OQ3 - PubMed Abstract:
Clostridium difficile is a burden to healthcare systems around the world, causing tens of thousands of deaths annually. The S-layer of the bacterium, a layer of protein found of the surface of cells, has received a significant amount of attention over the past two decades as a potential target to combat the growing threat presented by C. difficile infections. The S-layer contains a wide range of proteins, each of which possesses three cell wall-binding domains, while many also possess a "functional" region. Here, we present the high resolution structure of the functional region of one such protein, Cwp19 along with preliminary functional characterisation of the predicted glycoside hydrolase. Cwp19 has a TIM barrel fold and appears to possess a high degree of substrate selectivity. The protein also exhibits peptidoglycan hydrolase activity, an order of magnitude slower than that of lysozyme and is the first member of glycoside hydrolase-like family 10 to be characterised. This research goes some way to understanding the role of Cwp19 in the S-layer of C. difficile. Structural data are available in the PDB under the accession numbers 5OQ2 and 5OQ3.
- Department of Biology and Biochemistry, University of Bath, UK.
Organizational Affiliation: