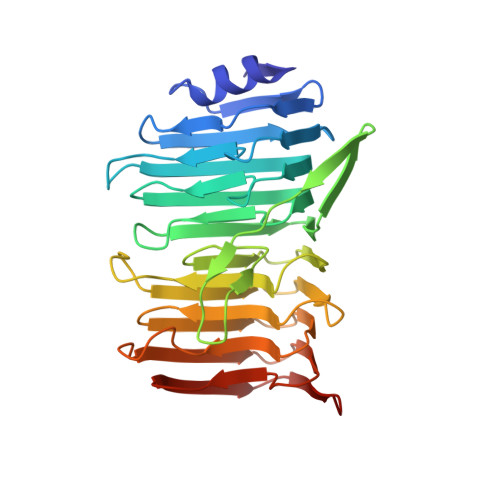
Structural basis for the role of serine-rich repeat proteins from Lactobacillus reuteriin gut microbe-host interactions.
Sequeira, S., Kavanaugh, D., MacKenzie, D.A., Suligoj, T., Walpole, S., Leclaire, C., Gunning, A.P., Latousakis, D., Willats, W.G.T., Angulo, J., Dong, C., Juge, N.(2018) Proc Natl Acad Sci U S A 115: E2706-E2715
- PubMed: 29507249
- DOI: https://doi.org/10.1073/pnas.1715016115
- Primary Citation of Related Structures:
5NXK, 5NY0 - PubMed Abstract:
Lactobacillus reuteri , a Gram-positive bacterial species inhabiting the gastrointestinal tract of vertebrates, displays remarkable host adaptation. Previous mutational analyses of rodent strain L. reuteri 100-23C identified a gene encoding a predicted surface-exposed serine-rich repeat protein (SRRP 100-23 ) that was vital for L. reuteri biofilm formation in mice. SRRPs have emerged as an important group of surface proteins on many pathogens, but no structural information is available in commensal bacteria. Here we report the 2.00-Å and 1.92-Å crystal structures of the binding regions (BRs) of SRRP 100-23 and SRRP 53608 from L. reuteri ATCC 53608, revealing a unique β-solenoid fold in this important adhesin family. SRRP 53608 -BR bound to host epithelial cells and DNA at neutral pH and recognized polygalacturonic acid (PGA), rhamnogalacturonan I, or chondroitin sulfate A at acidic pH. Mutagenesis confirmed the role of the BR putative binding site in the interaction of SRRP 53608 -BR with PGA. Long molecular dynamics simulations showed that SRRP 53608 -BR undergoes a pH-dependent conformational change. Together, these findings provide mechanistic insights into the role of SRRPs in host-microbe interactions and open avenues of research into the use of biofilm-forming probiotics against clinically important pathogens.
- BioMedical Research Centre, University of East Anglia, NR4 7TJ Norwich, United Kingdom.
Organizational Affiliation: