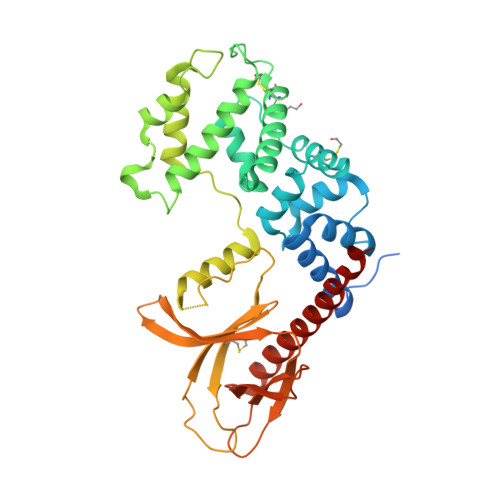
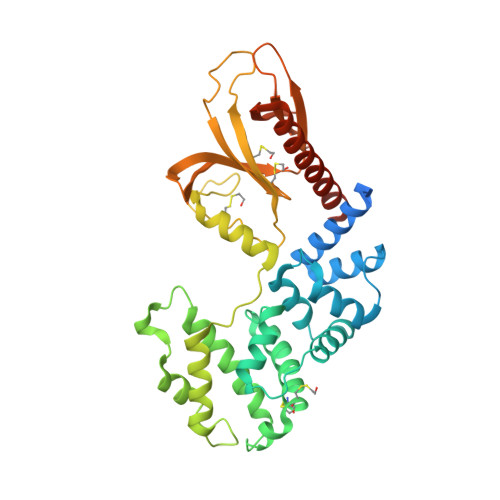
Multiple interactions between an Arf/GEF complex and charged lipids determine activation kinetics on the membrane.
Karandur, D., Nawrotek, A., Kuriyan, J., Cherfils, J.(2017) Proc Natl Acad Sci U S A 114: 11416-11421
- PubMed: 28923919
- DOI: https://doi.org/10.1073/pnas.1707970114
- Primary Citation of Related Structures:
5NLV, 5NLY - PubMed Abstract:
Lipidated small GTPases and their regulators need to bind to membranes to propagate actions in the cell, but an integrated understanding of how the lipid bilayer exerts its effect has remained elusive. Here we focused on ADP ribosylation factor (Arf) GTPases, which orchestrate a variety of regulatory functions in lipid and membrane trafficking, and their activation by the guanine-nucleotide exchange factor (GEF) Brag2, which controls integrin endocytosis and cell adhesion and is impaired in cancer and developmental diseases. Biochemical and structural data are available that showed the exceptional efficiency of Arf activation by Brag2 on membranes. We determined the high-resolution crystal structure of unbound Brag2 containing the GEF (Sec7) and membrane-binding (pleckstrin homology) domains, revealing that it has a constitutively active conformation. We used this structure to analyze the interaction of uncomplexed Brag2 and of the myristoylated Arf1/Brag2 complex with a phosphatidylinositol bisphosphate (PIP 2 ) -containing lipid bilayer, using coarse-grained molecular dynamics. These simulations revealed that the system forms a close-packed, oriented interaction with the membrane, in which multiple PIP 2 lipids bind the canonical lipid-binding site and unique peripheral sites of the PH domain, the Arf GTPase and, unexpectedly, the Sec7 domain. We cross-validated these predictions by reconstituting the binding and kinetics of Arf and Brag2 in artificial membranes. Our coarse-grained structural model thus suggests that the high efficiency of Brag2 requires interaction with multiple lipids and a well-defined orientation on the membrane, resulting in a local PIP 2 enrichment, which has the potential to signal toward the Arf pathway.
- Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720.
Organizational Affiliation: