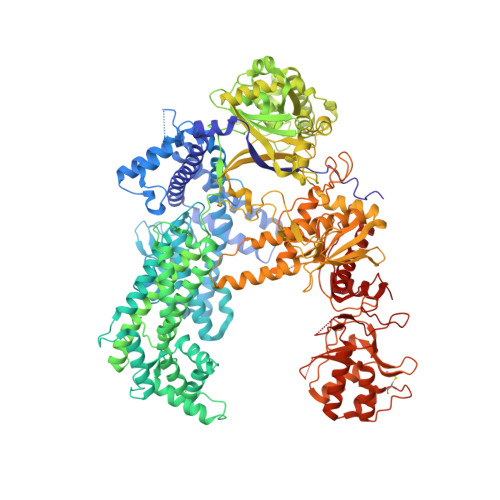
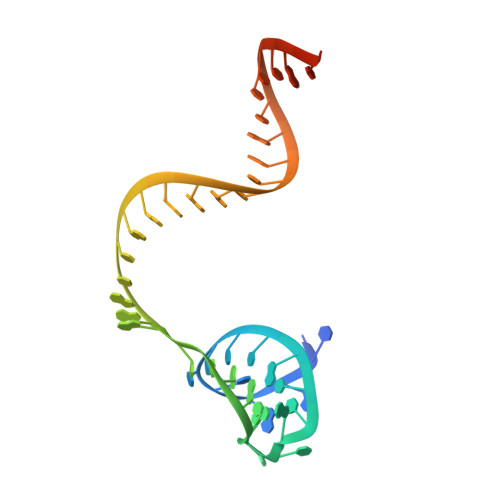
Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a.
Swarts, D.C., van der Oost, J., Jinek, M.(2017) Mol Cell 66: 221-233.e4
- PubMed: 28431230
- DOI: https://doi.org/10.1016/j.molcel.2017.03.016
- Primary Citation of Related Structures:
5NFV, 5NG6 - PubMed Abstract:
The CRISPR-associated protein Cas12a (Cpf1), which has been repurposed for genome editing, possesses two distinct nuclease activities: endoribonuclease activity for processing its own guide RNAs and RNA-guided DNase activity for target DNA cleavage. To elucidate the molecular basis of both activities, we determined crystal structures of Francisella novicida Cas12a bound to guide RNA and in complex with an R-loop formed by a non-cleavable guide RNA precursor and a full-length target DNA. Corroborated by biochemical experiments, these structures reveal the mechanisms of guide RNA processing and pre-ordering of the seed sequence in the guide RNA that primes Cas12a for target DNA binding. Furthermore, the R-loop complex structure reveals the strand displacement mechanism that facilitates guide-target hybridization and suggests a mechanism for double-stranded DNA cleavage involving a single active site. Together, these insights advance our mechanistic understanding of Cas12a enzymes and may contribute to further development of genome editing technologies.
- Department of Biochemistry, University of Zurich, CH-8057 Zurich, Switzerland.
Organizational Affiliation: