Kinetic and structural studies on the interactions of Torpedo californica acetylcholinesterase with two donepezil-like rigid analogues.
Caliandro, R., Pesaresi, A., Cariati, L., Procopio, A., Oliverio, M., Lamba, D.(2018) J Enzyme Inhib Med Chem 33: 794-803
- PubMed: 29651884
- DOI: https://doi.org/10.1080/14756366.2018.1458030
- Primary Citation of Related Structures:
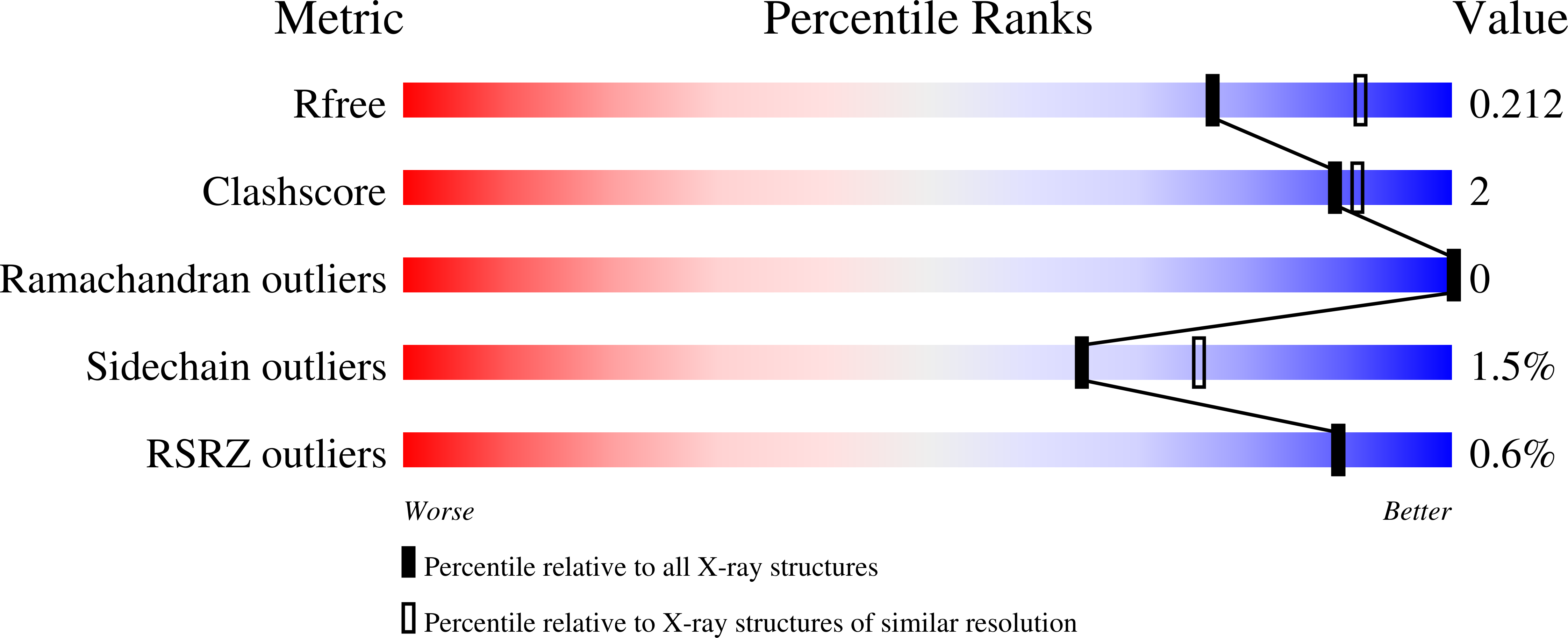
5NAP, 5NAU - PubMed Abstract:
Acetylcholinesterase inhibitors were introduced for the symptomatic treatment of Alzheimer's disease (AD). Among the currently approved inhibitors, donepezil (DNP) is one of the most preferred choices in AD therapy. The X-ray crystal structures of Torpedo californica AChE in complex with two novel rigid DNP-like analogs, compounds 1 and 2, have been determined. Kinetic studies indicated that compounds 1 and 2 show a mixed-type inhibition against TcAChE, with K i values of 11.12 ± 2.88 and 29.86 ± 1.12 nM, respectively. The DNP rigidification results in a likely entropy-enthalpy compensation with solvation effects contributing primarily to AChE binding affinity. Molecular docking evidenced the molecular basis for the binding of compounds 1 and 2 to the active site of β-secretase-1. Overall, these simplified DNP derivatives may represent new structural templates for the design of lead compounds for a more effective therapeutic strategy against AD by foreseeing a dual AChE and BACE-1 inhibitory activity.
- a Istituto di Cristallografia, Consiglio Nazionale delle Ricerche , Trieste , Italy.
Organizational Affiliation: