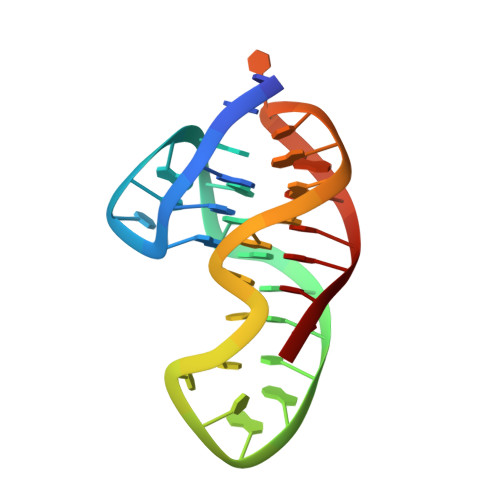
Solution-State Structure of a Long-Loop G-Quadruplex Formed Within Promoters of Plasmodium falciparum B var Genes.
Juribasic Kulcsar, M., Gabelica, V., Plavec, J.(2024) Chemistry 30: e202401190-e202401190
- PubMed: 38647110
- DOI: https://doi.org/10.1002/chem.202401190
- Primary Citation of Related Structures:
5MTA, 5MTG - PubMed Abstract:
We report the high-resolution NMR solution-state structure of an intramolecular G-quadruplex with a diagonal loop of ten nucleotides. The G-quadruplex is formed by a 34-nt DNA sequence, d[CAG 3 T 2 A 2 G 3 TATA 2 CT 3 AG 4 T 2 AG 3 T 2 ], named UpsB-Q-1. This sequence is found within promoters of the var genes of Plasmodium falciparum, which play a key role in malaria pathogenesis and evasion of the immune system. The [3+1]-hybrid G-quadruplex formed under physiologically relevant conditions exhibits a unique equilibrium between two structures, both stabilized by base stacking and non-canonical hydrogen bonding. Unique equilibrium of the two closely related 3D structures originates from a North-South repuckering of deoxyribose moiety of residue T27 in the lateral loop. Besides the 12 guanines involved in three G-quartets, most residues in loop regions are involved in interactions at both G-quartet-loop interfaces.
- Slovenian NMR Center, National Institute of Chemistry, Hajdrihova 19, SI-1000, Ljubljana, Slovenia.
Organizational Affiliation: