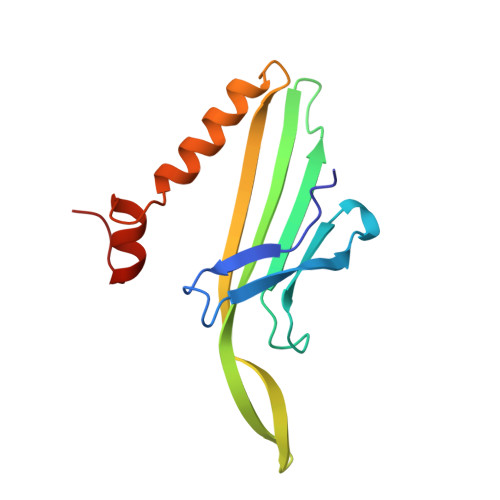
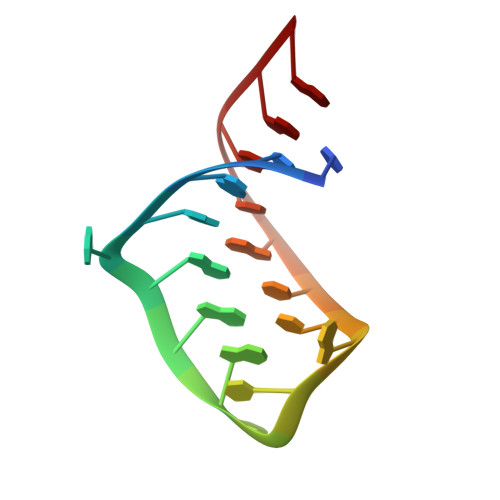
Crystal structures of a series of RNA aptamers complexed to the same protein target.
Rowsell, S., Stonehouse, N.J., Convery, M.A., Adams, C.J., Ellington, A.D., Hirao, I., Peabody, D.S., Stockley, P.G., Phillips, S.E.(1998) Nat Struct Biol 5: 970-975
- PubMed: 9808042
- DOI: https://doi.org/10.1038/2946
- Primary Citation of Related Structures:
5MSF, 7MSF - PubMed Abstract:
We have determined the crystal structures, at 2.8 A resolution, of two different RNA aptamers, each bound to MS2 coat protein. One of the aptamers contains a non-Watson-Crick base pair, while the other is missing one of the unpaired adenines that make sequence-specific contacts in the wild-type complex. Despite these differences, the RNA aptamers bind in the same location on the protein as the wild-type translational operator. Comparison of these new structures with other MS2-RNA complexes allows us to refine further the definition of the minimal recognition elements and suggests a possible application of the MS2 system for routine structure determination of small nucleic acid motifs.
- School of Biochemistry and Molecular Biology, and North of England Structural Biology Centre, University of Leeds, UK.
Organizational Affiliation: