Structure and biophysical characterization of the human full-length neurturin-GFRa2 complex: A role for heparan sulfate in signaling.
Sandmark, J., Dahl, G., Oster, L., Xu, B., Johansson, P., Akerud, T., Aagaard, A., Davidsson, P., Bigalke, J.M., Winzell, M.S., Rainey, G.J., Roth, R.G.(2018) J Biological Chem 293: 5492-5508
- PubMed: 29414779
- DOI: https://doi.org/10.1074/jbc.RA117.000820
- Primary Citation of Related Structures:
5MR4, 5MR5, 5NMZ - PubMed Abstract:
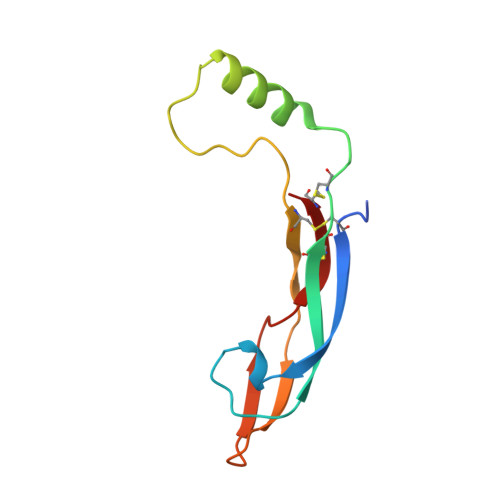
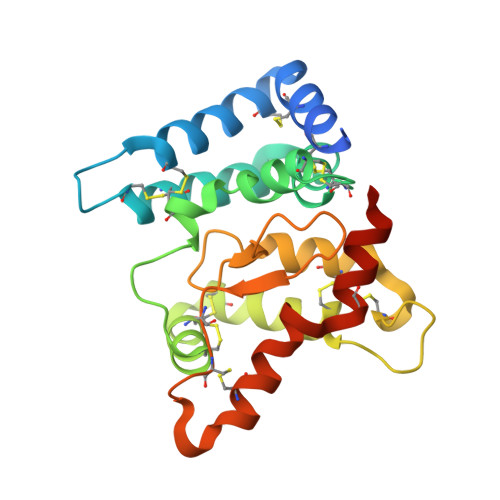
Neurturin (NRTN) provides trophic support to neurons and is considered a therapeutic agent for neurodegenerative diseases, such as Parkinson's disease. It binds to its co-receptor GFRa2, and the resulting NRTN-GFRa2 complex activates the transmembrane receptors rearranged during transfection (RET) or the neural cell adhesion molecule (NCAM). We report the crystal structure of NRTN, alone and in complex with GFRa2. This is the first crystal structure of a GFRa with all three domains and shows that domain 1 does not interact directly with NRTN, but it may support an interaction with RET and/or NCAM, via a highly conserved surface. In addition, biophysical results show that the relative concentration of GFRa2 on cell surfaces can affect the functional affinity of NRTN through avidity effects. We have identified a heparan sulfate-binding site on NRTN and a putative binding site in GFRa2, suggesting that heparan sulfate has a role in the assembly of the signaling complex. We further show that mutant NRTN with reduced affinity for heparan sulfate may provide a route forward for delivery of NRTN with increased exposure in preclinical in vivo models and ultimately to Parkinson's patients.
- From the Departments of Structure, Biophysics and Fragment-based Lead Generation, Discovery Sciences.
Organizational Affiliation: