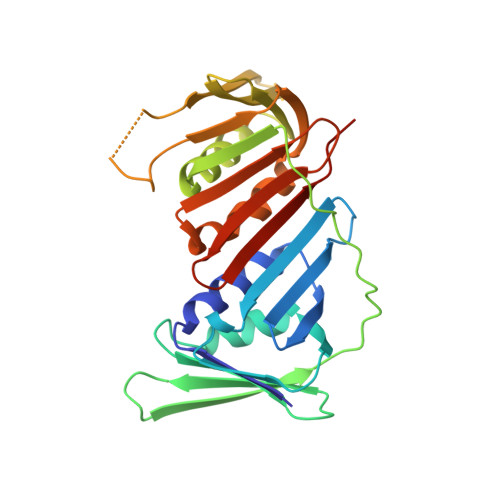
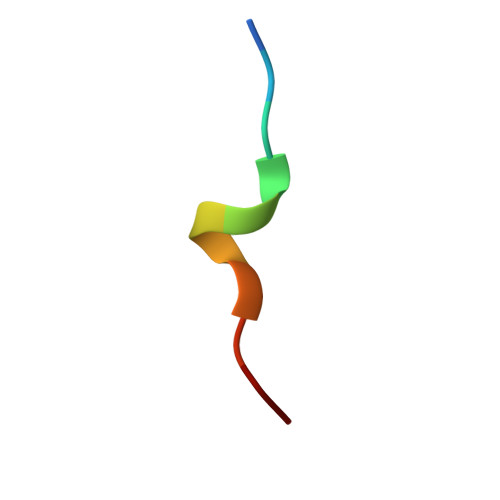Structural insights into the function of ZRANB3 in replication stress response.
Sebesta, M., Cooper, C.D.O., Ariza, A., Carnie, C.J., Ahel, D.(2017) Nat Commun 8: 15847-15847
- PubMed: 28621305
- DOI: https://doi.org/10.1038/ncomms15847
- Primary Citation of Related Structures:
5MKW, 5MLO, 5MLW - PubMed Abstract:
Strategies to resolve replication blocks are critical for the maintenance of genome stability. Among the factors implicated in the replication stress response is the ATP-dependent endonuclease ZRANB3. Here, we present the structure of the ZRANB3 HNH (His-Asn-His) endonuclease domain and provide a detailed analysis of its activity. We further define PCNA as a key regulator of ZRANB3 function, which recruits ZRANB3 to stalled replication forks and stimulates its endonuclease activity. Finally, we present the co-crystal structures of PCNA with two specific motifs in ZRANB3: the PIP box and the APIM motif. Our data provide important structural insights into the PCNA-APIM interaction, and reveal unexpected similarities between the PIP box and the APIM motif. We propose that PCNA and ATP-dependency serve as a multi-layered regulatory mechanism that modulates ZRANB3 activity at replication forks. Importantly, our findings allow us to interpret the functional significance of cancer associated ZRANB3 mutations.
- Sir William Dunn School of Pathology, University of Oxford, Oxford OX1 3RE, UK.
Organizational Affiliation: