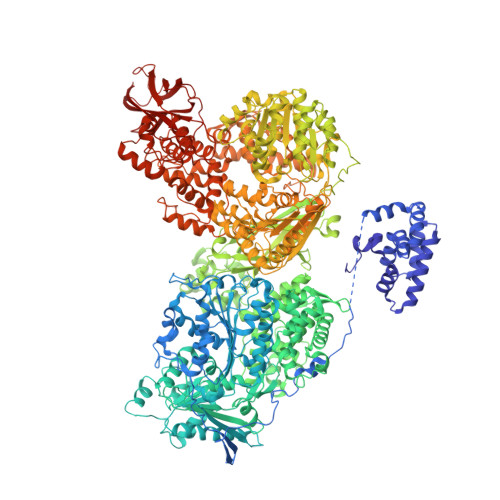
Interplay of cis- and trans-regulatory mechanisms in the spliceosomal RNA helicase Brr2.
Absmeier, E., Becke, C., Wollenhaupt, J., Santos, K.F., Wahl, M.C.(2017) Cell Cycle 16: 100-112
- PubMed: 27880071
- DOI: https://doi.org/10.1080/15384101.2016.1255384
- Primary Citation of Related Structures:
5M52, 5M59, 5M5P - PubMed Abstract:
RNA helicase Brr2 is implicated in multiple phases of pre-mRNA splicing and thus requires tight regulation. Brr2 can be auto-inhibited via a large N-terminal region folding back onto its helicase core and auto-activated by a catalytically inactive C-terminal helicase cassette. Furthermore, it can be regulated in trans by the Jab1 domain of the Prp8 protein, which can inhibit Brr2 by intermittently inserting a C-terminal tail in the enzyme's RNA-binding tunnel or activate the helicase after removal of this tail. Presently it is unclear, whether these regulatory mechanisms functionally interact and to which extent they are evolutionarily conserved. Here, we report crystal structures of Saccharomyces cerevisiae and Chaetomium thermophilum Brr2-Jab1 complexes, demonstrating that Jab1-based inhibition of Brr2 presumably takes effect in all eukaryotes but is implemented via organism-specific molecular contacts. Moreover, the structures show that Brr2 auto-inhibition can act in concert with Jab1-mediated inhibition, and suggest that the N-terminal region influences how the Jab1 C-terminal tail interacts at the RNA-binding tunnel. Systematic RNA binding and unwinding studies revealed that the N-terminal region and the Jab1 C-terminal tail specifically interfere with accommodation of double-stranded and single-stranded regions of an RNA substrate, respectively, mutually reinforcing each other. Additionally, such analyses show that regulation based on the N-terminal region requires the presence of the inactive C-terminal helicase cassette. Together, our results outline an intricate system of regulatory mechanisms, which control Brr2 activities during snRNP assembly and splicing.
- a Freie Universität Berlin, Laboratory of Structural Biochemistry , Berlin , Germany.
Organizational Affiliation: