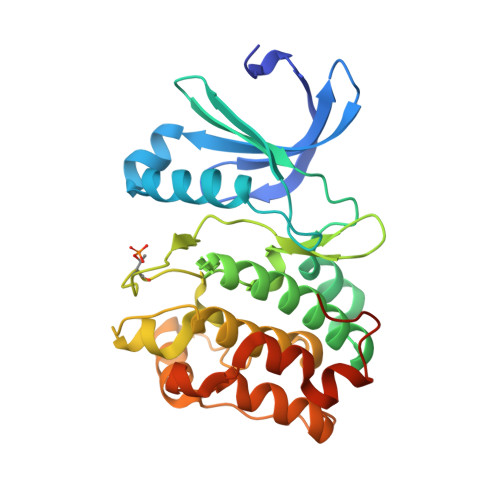
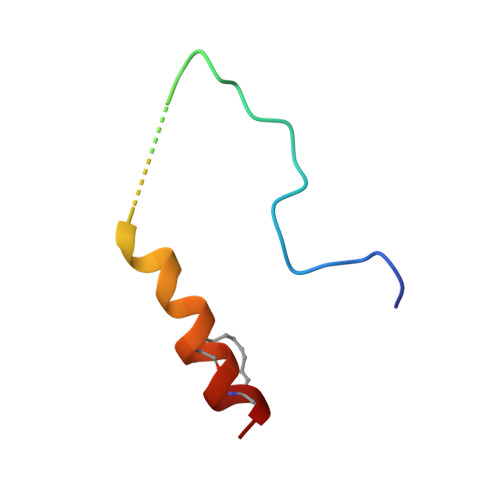
A TPX2 Proteomimetic Has Enhanced Affinity for Aurora-A Due to Hydrocarbon Stapling of a Helix.
Rennie, Y.K., McIntyre, P.J., Akindele, T., Bayliss, R., Jamieson, A.G.(2016) ACS Chem Biol 11: 3383-3390
- PubMed: 27775325
- DOI: https://doi.org/10.1021/acschembio.6b00727
- Primary Citation of Related Structures:
5LXM - PubMed Abstract:
Inhibition of protein kinases using ATP-competitive compounds is an important strategy in drug discovery. In contrast, the allosteric regulation of kinases through the disruption of protein-protein interactions has not been widely adopted, despite the potential for selective targeting. Aurora-A kinase regulates mitotic entry and mitotic spindle assembly and is a promising target for anticancer therapy. The microtubule-associated protein TPX2 activates Aurora-A through binding to two sites. Aurora-A recognition is mediated by two motifs within the first 43 residues of TPX2, connected by a flexible linker. To characterize the contributions of these three structural elements, we prepared a series of TPX2 proteomimetics and investigated their binding affinity for Aurora-A using isothermal titration calorimetry. A novel stapled TPX2 peptide was developed that has improved binding affinity for Aurora-A and mimics the function of TPX2 in activating Aurora-A's autophosphorylation. We conclude that the helical region of TPX2 folds upon binding Aurora-A, and that stabilization of this helix does not compromise Aurora-A activation. This study demonstrates that the preparation of these proteomimetics using modern synthesis methods is feasible and their biochemical evaluation demonstrates the power of proteomimetics as tool compounds for investigating PPIs involving intrinsically disordered regions of proteins.
- Department of Chemistry, University of Leicester , Lancaster Road, Leicester LE1 9HN, United Kingdom.
Organizational Affiliation: