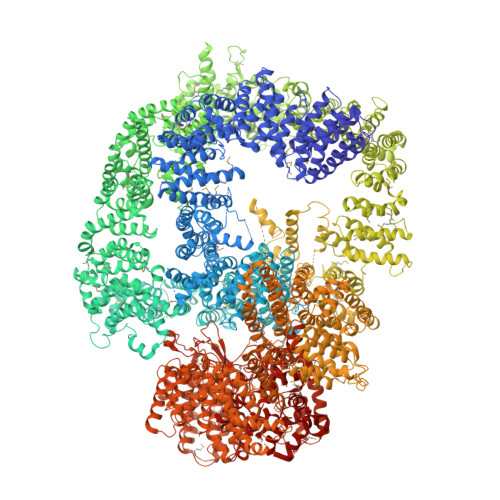
DNA-PKcs structure suggests an allosteric mechanism modulating DNA double-strand break repair.
Sibanda, B.L., Chirgadze, D.Y., Ascher, D.B., Blundell, T.L.(2017) Science 355: 520-524
- PubMed: 28154079
- DOI: https://doi.org/10.1126/science.aak9654
- Primary Citation of Related Structures:
5LUQ - PubMed Abstract:
DNA-dependent protein kinase catalytic subunit (DNA-PKcs) is a central component of nonhomologous end joining (NHEJ), repairing DNA double-strand breaks that would otherwise lead to apoptosis or cancer. We have solved its structure in complex with the C-terminal peptide of Ku80 at 4.3 angstrom resolution using x-ray crystallography. We show that the 4128-amino acid structure comprises three large structural units: the N-terminal unit, the Circular Cradle, and the Head. Conformational differences between the two molecules in the asymmetric unit are correlated with changes in accessibility of the kinase active site, which are consistent with an allosteric mechanism to bring about kinase activation. The location of KU80ct 194 in the vicinity of the breast cancer 1 (BRCA1) binding site suggests competition with BRCA1, leading to pathway selection between NHEJ and homologous recombination.
- Department of Biochemistry, University of Cambridge, Old Addenbrooke's Site, 80 Tennis Court Road, Cambridge CB2 1GA, UK.
Organizational Affiliation: