Identification of (R)-N-((4-Methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1-(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1H-indole-3-carboxamide (CPI-1205), a Potent and Selective Inhibitor of Histone Methyltransferase EZH2, Suitable for Phase I Clinical Trials for B-Cell Lymphomas.
Vaswani, R.G., Gehling, V.S., Dakin, L.A., Cook, A.S., Nasveschuk, C.G., Duplessis, M., Iyer, P., Balasubramanian, S., Zhao, F., Good, A.C., Campbell, R., Lee, C., Cantone, N., Cummings, R.T., Normant, E., Bellon, S.F., Albrecht, B.K., Harmange, J.C., Trojer, P., Audia, J.E., Zhang, Y., Justin, N., Chen, S., Wilson, J.R., Gamblin, S.J.(2016) J Med Chem 59: 9928-9941
- PubMed: 27739677
- DOI: https://doi.org/10.1021/acs.jmedchem.6b01315
- Primary Citation of Related Structures:
5LS6 - PubMed Abstract:
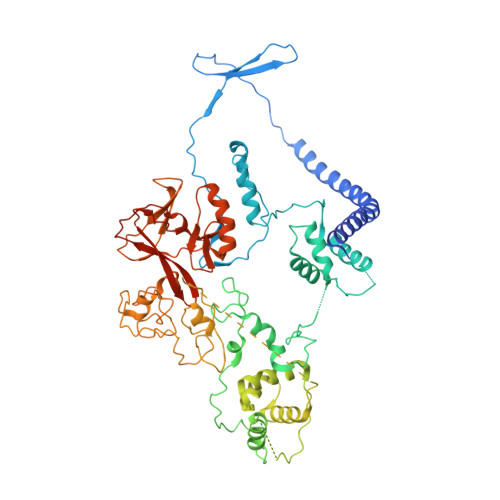
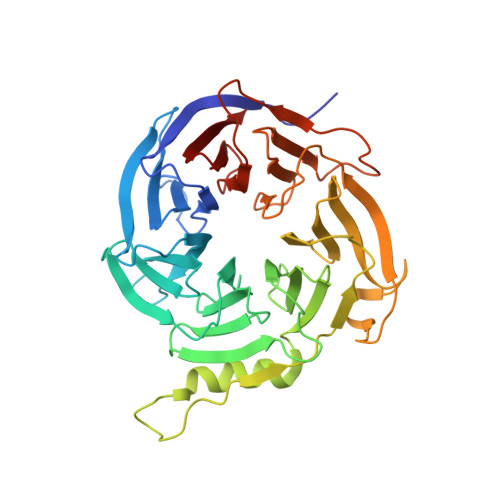
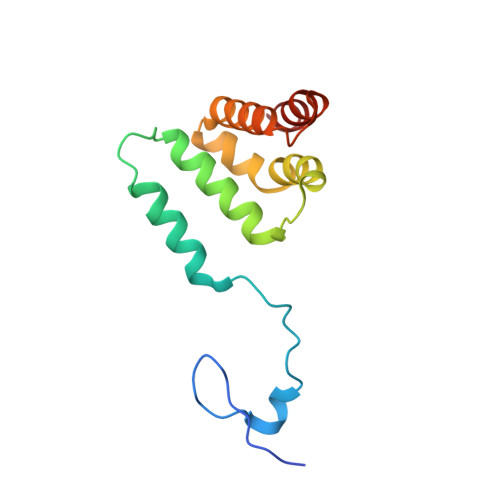
Polycomb repressive complex 2 (PRC2) has been shown to play a major role in transcriptional silencing in part by installing methylation marks on lysine 27 of histone 3. Dysregulation of PRC2 function correlates with certain malignancies and poor prognosis. EZH2 is the catalytic engine of the PRC2 complex and thus represents a key candidate oncology target for pharmacological intervention. Here we report the optimization of our indole-based EZH2 inhibitor series that led to the identification of CPI-1205, a highly potent (biochemical IC 50 = 0.002 μM, cellular EC 50 = 0.032 μM) and selective inhibitor of EZH2. This compound demonstrates robust antitumor effects in a Karpas-422 xenograft model when dosed at 160 mg/kg BID and is currently in Phase I clinical trials. Additionally, we disclose the co-crystal structure of our inhibitor series bound to the human PRC2 complex.
- Constellation Pharmaceuticals, Inc., 215 First Street, Suite 200, Cambridge, Massachusetts 02142, United States.
Organizational Affiliation: