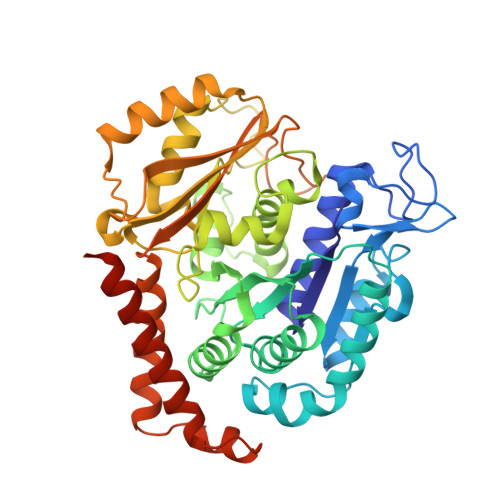
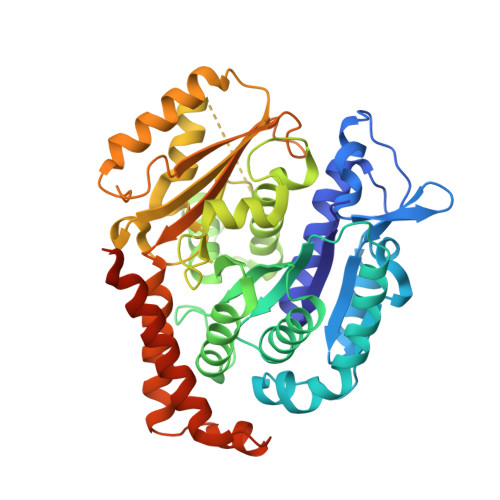
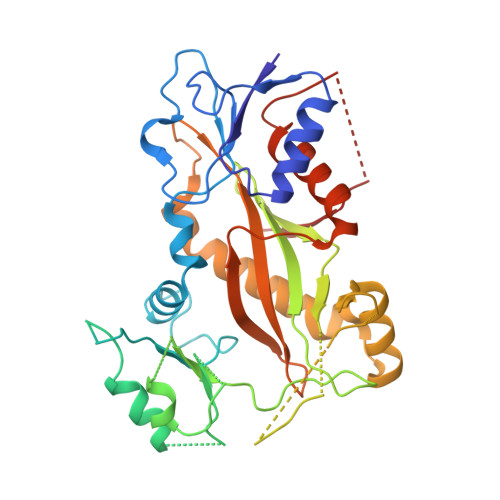
The synthetic diazonamide DZ-2384 has distinct effects on microtubule curvature and dynamics without neurotoxicity.
Wieczorek, M., Tcherkezian, J., Bernier, C., Prota, A.E., Chaaban, S., Rolland, Y., Godbout, C., Hancock, M.A., Arezzo, J.C., Ocal, O., Rocha, C., Olieric, N., Hall, A., Ding, H., Bramoulle, A., Annis, M.G., Zogopoulos, G., Harran, P.G., Wilkie, T.M., Brekken, R.A., Siegel, P.M., Steinmetz, M.O., Shore, G.C., Brouhard, G.J., Roulston, A.(2016) Sci Transl Med 8: 365ra159-365ra159
- PubMed: 27856798
- DOI: https://doi.org/10.1126/scitranslmed.aag1093
- Primary Citation of Related Structures:
5LOV - PubMed Abstract:
Microtubule-targeting agents (MTAs) are widely used anticancer agents, but toxicities such as neuropathy limit their clinical use. MTAs bind to and alter the stability of microtubules, causing cell death in mitosis. We describe DZ-2384, a preclinical compound that exhibits potent antitumor activity in models of multiple cancer types. It has an unusually high safety margin and lacks neurotoxicity in rats at effective plasma concentrations. DZ-2384 binds the vinca domain of tubulin in a distinct way, imparting structurally and functionally different effects on microtubule dynamics compared to other vinca-binding compounds. X-ray crystallography and electron microscopy studies demonstrate that DZ-2384 causes straightening of curved protofilaments, an effect proposed to favor polymerization of tubulin. Both DZ-2384 and the vinca alkaloid vinorelbine inhibit microtubule growth rate; however, DZ-2384 increases the rescue frequency and preserves the microtubule network in nonmitotic cells and in primary neurons. This differential modulation of tubulin results in a potent MTA therapeutic with enhanced safety.
- Department of Biology, McGill University, Montreal, Quebec H3A 1B1, Canada.
Organizational Affiliation: