Protein Surface Topography as a tool to enhance the selective activity of a potassium channel blocker.
Berkut, A.A., Chugunov, A.O., Mineev, K.S., Peigneur, S., Tabakmakher, V.M., Krylov, N.A., Oparin, P.B., Lihonosova, A.F., Novikova, E.V., Arseniev, A.S., Grishin, E.V., Tytgat, J., Efremov, R.G., Vassilevski, A.A.(2019) J Biological Chem
- PubMed: 31533989
- DOI: https://doi.org/10.1074/jbc.RA119.010494
- Primary Citation of Related Structures:
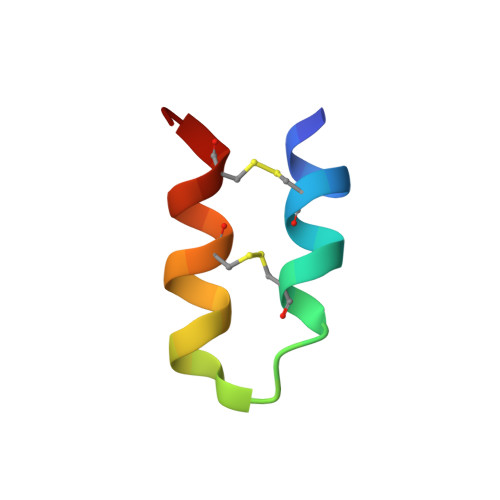
5LM0 - PubMed Abstract:
Tk-hefu is an artificial peptide designed based on the α-hairpinin scaffold, which selectively blocks voltage-gated potassium channels K v 1.3. Here we present its spatial structure resolved by NMR spectroscopy and analyze its interaction with channels using computer modeling. We apply protein surface topography to suggest mutations and increase Tk-hefu affinity to the K v 1.3 channel isoform. We redesign the functional surface of Tk-hefu to better match the respective surface of the channel pore vestibule. The resulting peptide Tk-hefu-2 retains K v 1.3 selectivity and displays ∼15 times greater activity compared with Tk-hefu. We verify the mode of Tk-hefu-2 binding to the channel outer vestibule experimentally by site-directed mutagenesis. We argue that scaffold engineering aided by protein surface topography represents a reliable tool for design and optimization of specific ion channel ligands.
- M.M. Shemyakin & Yu.A. Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, 117997 Moscow, Russia.
Organizational Affiliation: