Guanine Can Direct Binding Specificity of Ru-dipyridophenazine (dppz) Complexes to DNA through Steric Effects.
Hall, J.P., Gurung, S.P., Henle, J., Poidl, P., Andersson, J., Lincoln, P., Winter, G., Sorensen, T., Cardin, D.J., Brazier, J.A., Cardin, C.J.(2017) Chemistry 23: 4981-4985
- PubMed: 28105682
- DOI: https://doi.org/10.1002/chem.201605508
- Primary Citation of Related Structures:
5LFS, 5LFW, 5LFX - PubMed Abstract:
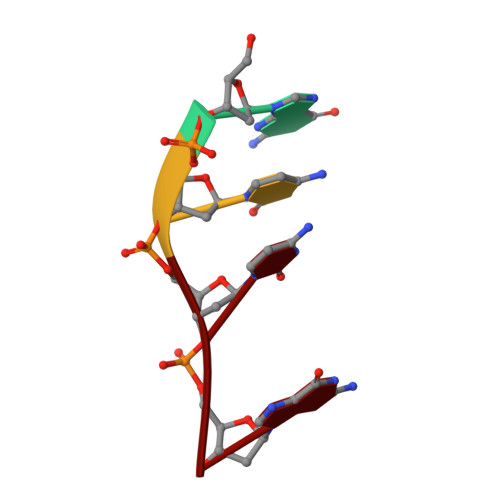
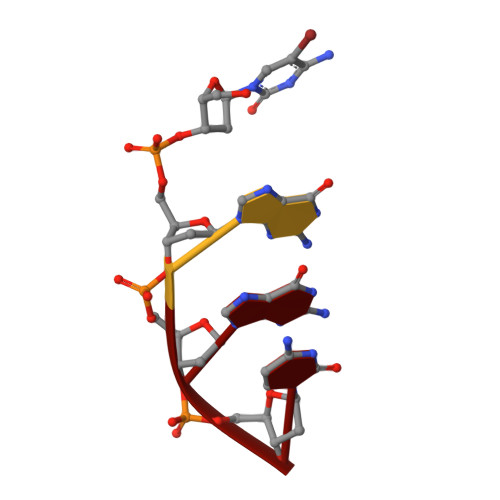
X-ray crystal structures of three Λ-[Ru(L) 2 dppz] 2+ complexes (dppz=dipyridophenazine; L=1,10-phenanthroline (phen), 2,2'-bipyridine (bpy)) bound to d((5BrC)GGC/GCCG) showed the compounds intercalated at a 5'-CG-3' step. The compounds bind through canted intercalation, with the binding angle determined by the guanine NH 2 group, in contrast to symmetrical intercalation previously observed at 5'-TA-3' sites. This result suggests that canted intercalation is preferred at 5'-CG-3' sites even though the site itself is symmetrical, and we hypothesise that symmetrical intercalation in a 5'-CG-3' step could give rise to a longer luminescence lifetime than canted intercalation.
- Department of Chemistry, University of Reading, Whiteknights, Reading, RG6 6AD, UK.
Organizational Affiliation: