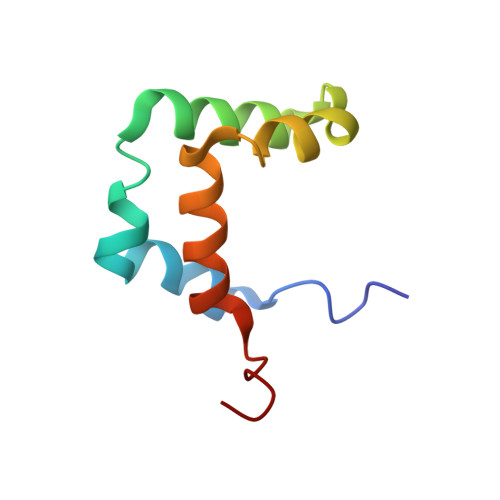
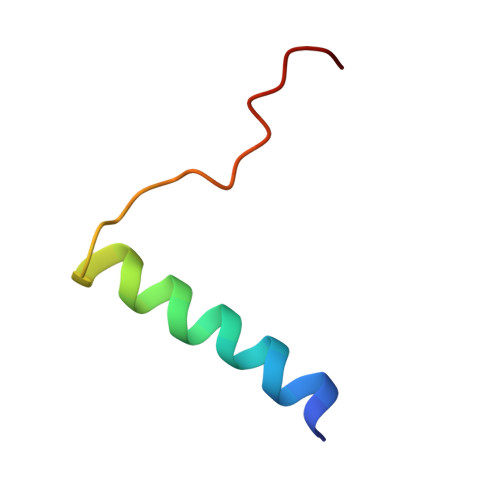
Structural Features of the Box C/D snoRNP Pre-assembly Process Are Conserved through Species.
Quinternet, M., Chagot, M.E., Rothe, B., Tiotiu, D., Charpentier, B., Manival, X.(2016) Structure 24: 1693-1706
- PubMed: 27594683
- DOI: https://doi.org/10.1016/j.str.2016.07.016
- Primary Citation of Related Structures:
5L85 - PubMed Abstract:
Box C/D small nucleolar ribonucleoparticles (snoRNPs) support 2'-O-methylation of several target RNAs. They share a common set of four core proteins (SNU13, NOP58, NOP56, and FBL) that are assembled on different guide small nucleolar RNAs. Assembly of these entities involves additional protein factors that are absent in the mature active particle. In this context, the platform protein NUFIP1/Rsa1 establishes direct and simultaneous contacts with core proteins and with the components of the assembly machinery. Here, we solve the nuclear magnetic resonance (NMR) structure of a complex resulting from interaction between protein fragments of human NUFIP1 and its cofactor ZNHIT3, and emphasize their imbrication. Using yeast two-hybrid and complementation assays, protein co-expression, isothermal titration calorimetry, and NMR, we demonstrate that yeast and human complexes involving NUFIP1/Rsa1p, ZNHIT3/Hit1p, and SNU13/Snu13p share strong structural similarities, suggesting that the initial steps of the box C/D snoRNP assembly process are conserved among species.
- FR CNRS-3209 Bioingénierie Moléculaire, Cellulaire et Thérapeutique (BMCT), CNRS, Université de Lorraine, Biopôle, Campus Biologie-Santé, CS 50184, 54505 Vandœuvre-lès-Nancy Cedex, France.
Organizational Affiliation: