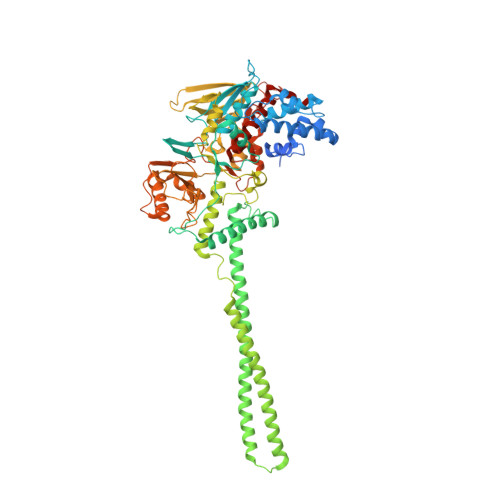
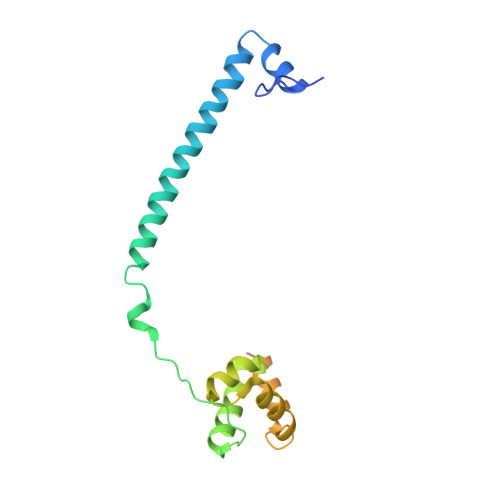
Polymyxins and quinazolines are LSD1/KDM1A inhibitors with unusual structural features.
Speranzini, V., Rotili, D., Ciossani, G., Pilotto, S., Marrocco, B., Forgione, M., Lucidi, A., Forneris, F., Mehdipour, P., Velankar, S., Mai, A., Mattevi, A.(2016) Sci Adv 2: e1601017-e1601017
- PubMed: 27626075
- DOI: https://doi.org/10.1126/sciadv.1601017
- Primary Citation of Related Structures:
5L3E, 5L3F, 5L3G, 5LBQ - PubMed Abstract:
Because of its involvement in the progression of several malignant tumors, the histone lysine-specific demethylase 1 (LSD1) has become a prominent drug target in modern medicinal chemistry research. We report on the discovery of two classes of noncovalent inhibitors displaying unique structural features. The antibiotics polymyxins bind at the entrance of the substrate cleft, where their highly charged cyclic moiety interacts with a cluster of positively charged amino acids. The same site is occupied by quinazoline-based compounds, which were found to inhibit the enzyme through a most peculiar mode because they form a pile of five to seven molecules that obstruct access to the active center. These data significantly indicate unpredictable strategies for the development of epigenetic inhibitors.
- Department of Biology and Biotechnology, University of Pavia, 27100 Pavia, Italy.
Organizational Affiliation: