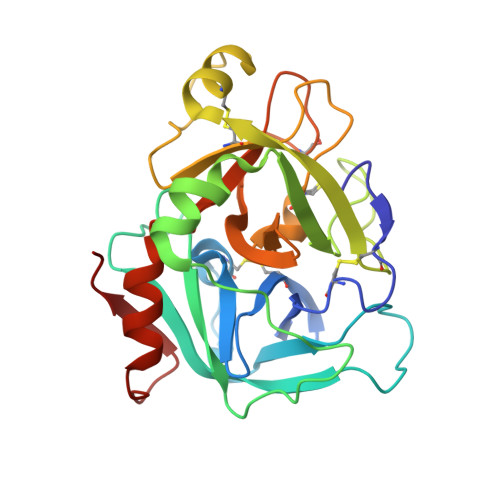
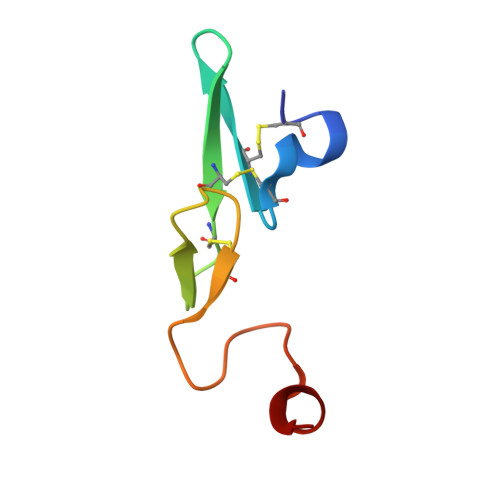
Synthesis and P1' SAR exploration of potent macrocyclic tissue factor-factor VIIa inhibitors.
Ladziata, V.U., Glunz, P.W., Zou, Y., Zhang, X., Jiang, W., Jacutin-Porte, S., Cheney, D.L., Wei, A., Luettgen, J.M., Harper, T.M., Wong, P.C., Seiffert, D., Wexler, R.R., Priestley, E.S.(2016) Bioorg Med Chem Lett 26: 5051-5057
- PubMed: 27612545
- DOI: https://doi.org/10.1016/j.bmcl.2016.08.088
- Primary Citation of Related Structures:
5L2Y, 5L2Z, 5L30 - PubMed Abstract:
Selective tissue factor-factor VIIa complex (TF-FVIIa) inhibitors are viewed as promising compounds for treating thrombotic disease. In this contribution, we describe multifaceted exploratory SAR studies of S1'-binding moieties within a macrocyclic chemotype aimed at replacing cyclopropyl sulfone P1' group. Over the course of the optimization efforts, the 1-(1H-tetrazol-5-yl)cyclopropane P1' substituent emerged as an improved alternative, offering increased metabolic stability and lower clearance, while maintaining excellent potency and selectivity.
- Bristol-Myers Squibb Research & Development, Princeton, NJ 08543, United States. Electronic address: vladimir.ladziata@bms.com.
Organizational Affiliation: