A Structural Investigation into Oct4 Regulation by Orphan Nuclear Receptors, Germ Cell Nuclear Factor (GCNF), and Liver Receptor Homolog-1 (LRH-1).
Weikum, E.R., Tuntland, M.L., Murphy, M.N., Ortlund, E.A.(2016) J Mol Biology 428: 4981-4992
- PubMed: 27984042
- DOI: https://doi.org/10.1016/j.jmb.2016.10.025
- Primary Citation of Related Structures:
5KRB, 5L0M - PubMed Abstract:
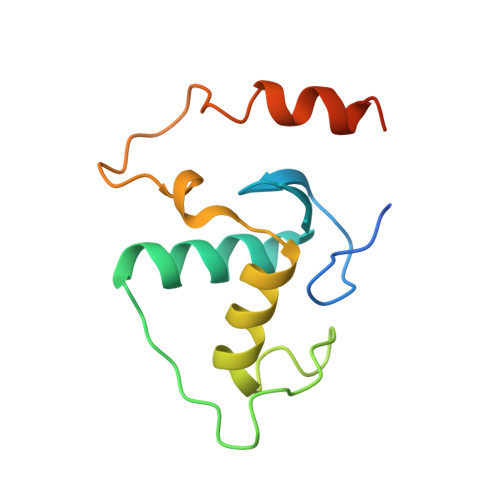
Oct4 is a transcription factor required for maintaining pluripotency and self-renewal in stem cells. Prior to differentiation, Oct4 must be silenced to allow for the development of the three germ layers in the developing embryo. This fine-tuning is controlled by the nuclear receptors (NRs), liver receptor homolog-1 (LRH-1) and germ cell nuclear factor (GCNF). Liver receptor homolog-1 is responsible for driving the expression of Oct4 where GCNF represses its expression upon differentiation. Both receptors bind to a DR0 motif located within the Oct4 promoter. Here, we present the first structure of mouse GCNF DNA-binding domain in complex with the Oct4 DR0. The overall structure revealed two molecules bound in a head-to-tail fashion on opposite sides of the DNA. Additionally, we solved the structure of the human LRH-1 DNA-binding domain bound to the same element. We explore the structural elements that govern Oct4 recognition by these two NRs.
- Department of Biochemistry, Emory University School of Medicine, Atlanta, GA, 30322 USA.
Organizational Affiliation: