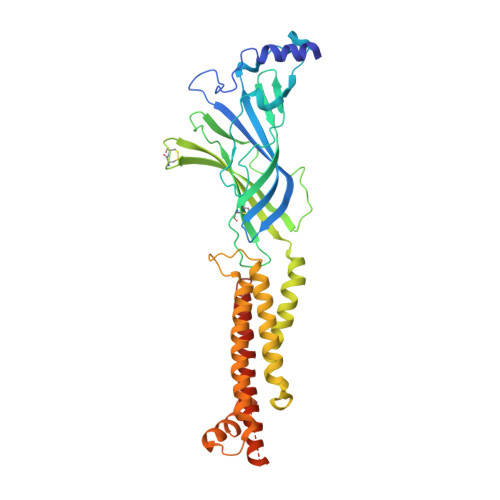
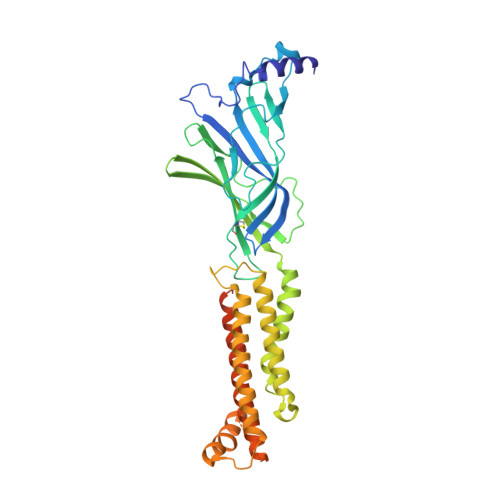
X-ray structure of the human alpha 4 beta 2 nicotinic receptor.
Morales-Perez, C.L., Noviello, C.M., Hibbs, R.E.(2016) Nature 538: 411-415
- PubMed: 27698419
- DOI: https://doi.org/10.1038/nature19785
- Primary Citation of Related Structures:
5KXI - PubMed Abstract:
Nicotinic acetylcholine receptors are ligand-gated ion channels that mediate fast chemical neurotransmission at the neuromuscular junction and have diverse signalling roles in the central nervous system. The nicotinic receptor has been a model system for cell-surface receptors, and specifically for ligand-gated ion channels, for well over a century. In addition to the receptors' prominent roles in the development of the fields of pharmacology and neurobiology, nicotinic receptors are important therapeutic targets for neuromuscular disease, addiction, epilepsy and for neuromuscular blocking agents used during surgery. The overall architecture of the receptor was described in landmark studies of the nicotinic receptor isolated from the electric organ of Torpedo marmorata. Structures of a soluble ligand-binding domain have provided atomic-scale insights into receptor-ligand interactions, while high-resolution structures of other members of the pentameric receptor superfamily provide touchstones for an emerging allosteric gating mechanism. All available high-resolution structures are of homopentameric receptors. However, the vast majority of pentameric receptors (called Cys-loop receptors in eukaryotes) present physiologically are heteromeric. Here we present the X-ray crystallographic structure of the human α4β2 nicotinic receptor, the most abundant nicotinic subtype in the brain. This structure provides insights into the architectural principles governing ligand recognition, heteromer assembly, ion permeation and desensitization in this prototypical receptor class.
- Departments of Neuroscience and Biophysics, University of Texas Southwestern Medical Center, Dallas, Texas 75390, USA.
Organizational Affiliation: