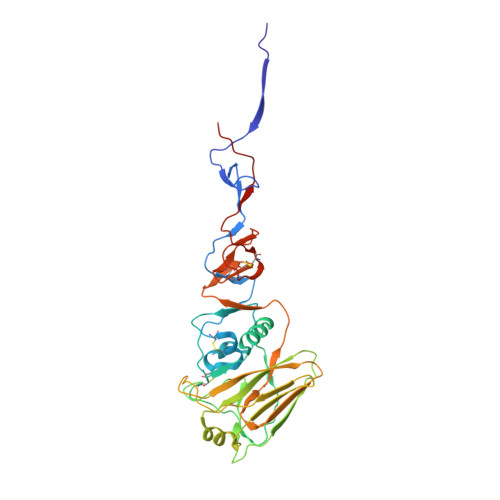
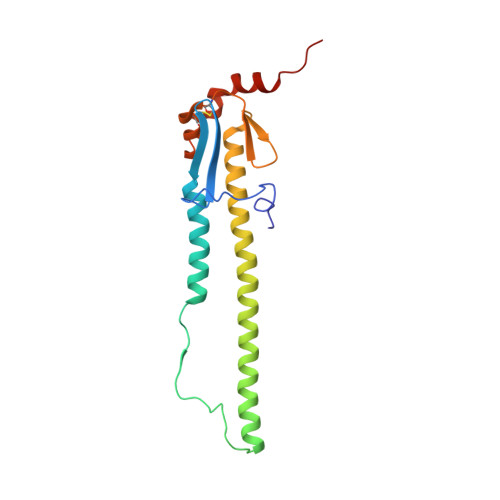
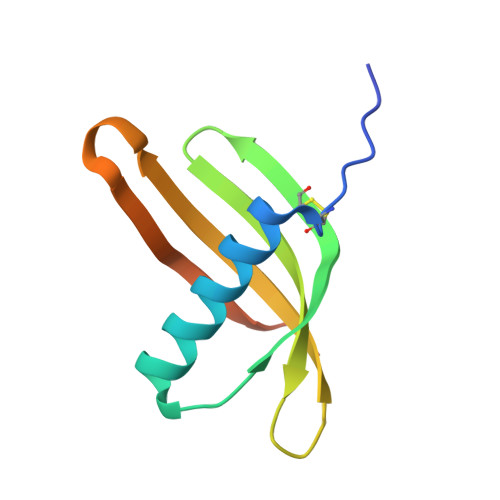
Computational design of trimeric influenza-neutralizing proteins targeting the hemagglutinin receptor binding site.
Strauch, E.M., Bernard, S.M., La, D., Bohn, A.J., Lee, P.S., Anderson, C.E., Nieusma, T., Holstein, C.A., Garcia, N.K., Hooper, K.A., Ravichandran, R., Nelson, J.W., Sheffler, W., Bloom, J.D., Lee, K.K., Ward, A.B., Yager, P., Fuller, D.H., Wilson, I.A., Baker, D.(2017) Nat Biotechnol 35: 667-671
- PubMed: 28604661
- DOI: https://doi.org/10.1038/nbt.3907
- Primary Citation of Related Structures:
5KUX, 5KUY - PubMed Abstract:
Many viral surface glycoproteins and cell surface receptors are homo-oligomers, and thus can potentially be targeted by geometrically matched homo-oligomers that engage all subunits simultaneously to attain high avidity and/or lock subunits together. The adaptive immune system cannot generally employ this strategy since the individual antibody binding sites are not arranged with appropriate geometry to simultaneously engage multiple sites in a single target homo-oligomer. We describe a general strategy for the computational design of homo-oligomeric protein assemblies with binding functionality precisely matched to homo-oligomeric target sites. In the first step, a small protein is designed that binds a single site on the target. In the second step, the designed protein is assembled into a homo-oligomer such that the designed binding sites are aligned with the target sites. We use this approach to design high-avidity trimeric proteins that bind influenza A hemagglutinin (HA) at its conserved receptor binding site. The designed trimers can both capture and detect HA in a paper-based diagnostic format, neutralizes influenza in cell culture, and completely protects mice when given as a single dose 24 h before or after challenge with influenza.
- Department of Biochemistry, University of Washington, Seattle, Washington, USA.
Organizational Affiliation: