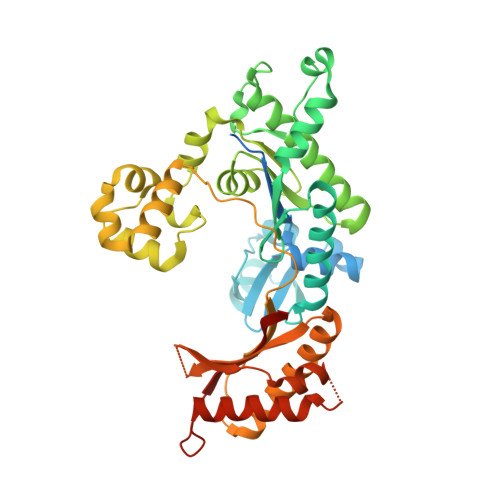
Kinetic and Structural Impact of Metal Ions and Genetic Variations on Human DNA Polymerase iota.
Choi, J.Y., Patra, A., Yeom, M., Lee, Y.S., Zhang, Q., Egli, M., Guengerich, F.P.(2016) J Biological Chem 291: 21063-21073
- PubMed: 27555320
- DOI: https://doi.org/10.1074/jbc.M116.748285
- Primary Citation of Related Structures:
5KT2, 5KT3, 5KT4, 5KT5, 5KT6, 5KT7 - PubMed Abstract:
DNA polymerase (pol) ι is a Y-family polymerase involved in translesion synthesis, exhibiting higher catalytic activity with Mn 2+ than Mg 2+ The human germline R96G variant impairs both Mn 2+ -dependent and Mg 2+ -dependent activities of pol ι, whereas the Δ1-25 variant selectively enhances its Mg 2+ -dependent activity. We analyzed pre-steady-state kinetic and structural effects of these two metal ions and genetic variations on pol ι using pol ι core (residues 1-445) proteins. The presence of Mn 2+ (0.15 mm) instead of Mg 2+ (2 mm) caused a 770-fold increase in efficiency (k pol /K d ,dCTP ) of pol ι for dCTP insertion opposite G, mainly due to a 450-fold decrease in K d ,dCTP The R96G and Δ1-25 variants displayed a 53-fold decrease and a 3-fold increase, respectively, in k pol /K d ,dCTP for dCTP insertion opposite G with Mg 2+ when compared with wild type, substantially attenuated by substitution with Mn 2+ Crystal structures of pol ι ternary complexes, including the primer terminus 3'-OH and a non-hydrolyzable dCTP analogue opposite G with the active-site Mg 2+ or Mn 2+ , revealed that Mn 2+ achieves more optimal octahedral coordination geometry than Mg 2+ , with lower values in average coordination distance geometry in the catalytic metal A-site. Crystal structures of R96G revealed the loss of three H-bonds of residues Gly-96 and Tyr-93 with an incoming dNTP, due to the lack of an arginine, as well as a destabilized Tyr-93 side chain secondary to the loss of a cation-π interaction between both side chains. These results provide a mechanistic basis for alteration in pol ι catalytic function with coordinating metals and genetic variation.
- From the Division of Pharmacology, Department of Molecular Cell Biology, Samsung Biomedical Research Institute, Sungkyunkwan University School of Medicine, Gyeonggi-do 16419, Republic of Korea.
Organizational Affiliation: