Crystal Structure of a Possible Enoyl-(acyl-carrier-protein) Reductase from Brucella melitensis
Dranow, D.M., Delker, S.L., Lorimer, D.D., Edwards, T.E.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Enoyl-[acyl-carrier-protein] reductase [NADH] | 293 | Brucella abortus bv. 1 str. 9-941 | Mutation(s): 0 Gene Names: fabI-2, BruAb1_2144 EC: 1.3.1.9 | 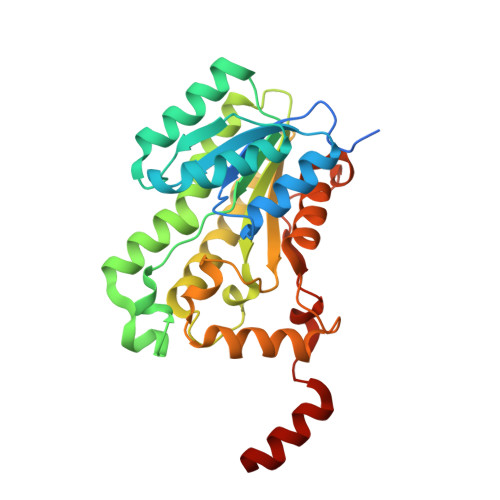 | |
UniProt | |||||
Find proteins for Q57A95 (Brucella abortus biovar 1 (strain 9-941)) Explore Q57A95 Go to UniProtKB: Q57A95 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q57A95 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAD Query on NAD | I [auth A] J [auth B] K [auth C] L [auth D] M [auth E] | NICOTINAMIDE-ADENINE-DINUCLEOTIDE C21 H27 N7 O14 P2 BAWFJGJZGIEFAR-NNYOXOHSSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 64.91 | α = 90.25 |
| b = 83.83 | β = 100.08 |
| c = 106.29 | γ = 90.82 |
| Software Name | Purpose |
|---|---|
| XSCALE | data scaling |
| PHASER | phasing |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |