Response to Comment on "Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2".
Jiao, L., Liu, X.(2016) Science 354: 1543-1543
- PubMed: 28008038
- DOI: https://doi.org/10.1126/science.aaj2335
- Primary Citation of Related Structures:
5KJH, 5KJI, 5KKL - PubMed Abstract:
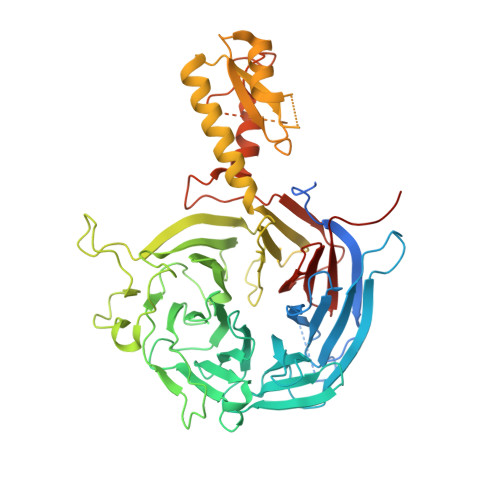
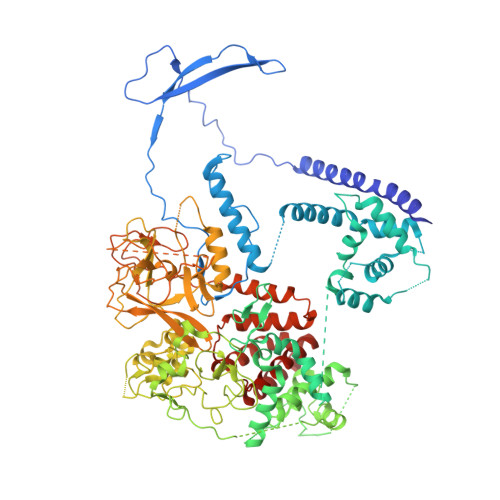
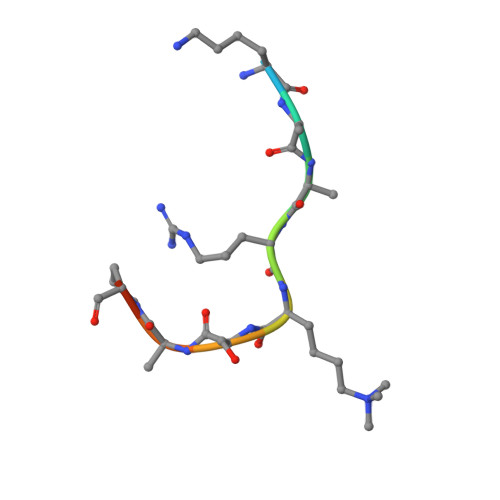
Zhang et al suggested that in the crystal structure of a polycomb repressive complex 2 from Chaetomium thermophilum (ctPRC2), a flexible linker region, but not the H3K27M cancer mutant peptide, better fits the electron density. Based on our new data, we agree with this alternative interpretation and provide the crystal structure of ctPRC2 bound to a bona fide H3K27M sequence.
- Cecil H. and Ida Green Center for Reproductive Biology Sciences and Division of Basic Research, Department of Obstetrics and Gynecology and Department of Biophysics, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA.
Organizational Affiliation: