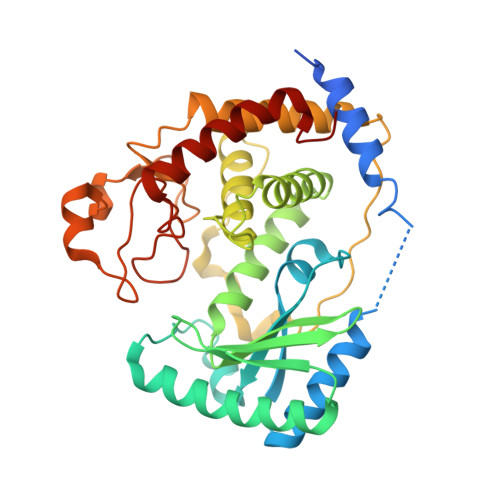
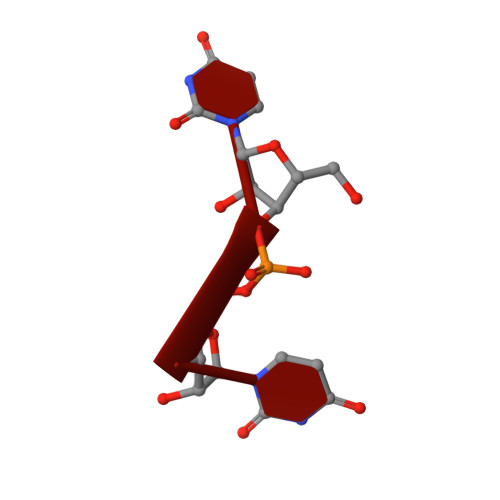
RNA Editing TUTase 1: structural foundation of substrate recognition, complex interactions and drug targeting.
Rajappa-Titu, L., Suematsu, T., Munoz-Tello, P., Long, M., Demir, O., Cheng, K.J., Stagno, J.R., Luecke, H., Amaro, R.E., Aphasizheva, I., Aphasizhev, R., Thore, S.(2016) Nucleic Acids Res 44: 10862-10878
- PubMed: 27744351
- DOI: https://doi.org/10.1093/nar/gkw917
- Primary Citation of Related Structures:
5HZD, 5I49, 5IDO, 5KAL - PubMed Abstract:
Terminal uridyltransferases (TUTases) execute 3' RNA uridylation across protists, fungi, metazoan and plant species. Uridylation plays a particularly prominent role in RNA processing pathways of kinetoplastid protists typified by the causative agent of African sleeping sickness, Trypanosoma brucei In mitochondria of this pathogen, most mRNAs are internally modified by U-insertion/deletion editing while guide RNAs and rRNAs are U-tailed. The founding member of TUTase family, RNA editing TUTase 1 (RET1), functions as a subunit of the 3' processome in uridylation of gRNA precursors and mature guide RNAs. Along with KPAP1 poly(A) polymerase, RET1 also participates in mRNA translational activation. RET1 is divergent from human TUTases and is essential for parasite viability in the mammalian host and the insect vector. Given its robust in vitro activity, RET1 represents an attractive target for trypanocide development. Here, we report high-resolution crystal structures of the RET1 catalytic core alone and in complex with UTP analogs. These structures reveal a tight docking of the conserved nucleotidyl transferase bi-domain module with a RET1-specific C2H2 zinc finger and RNA recognition (RRM) domains. Furthermore, we define RET1 region required for incorporation into the 3' processome, determinants for RNA binding, subunit oligomerization and processive UTP incorporation, and predict druggable pockets.
- Department of Molecular Biology, University of Geneva, 1211 Geneva, Switzerland.
Organizational Affiliation: