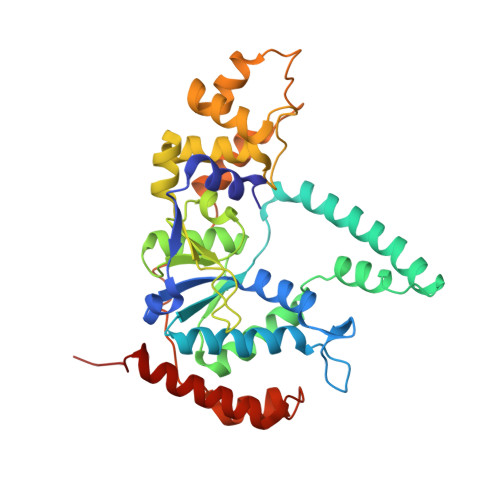
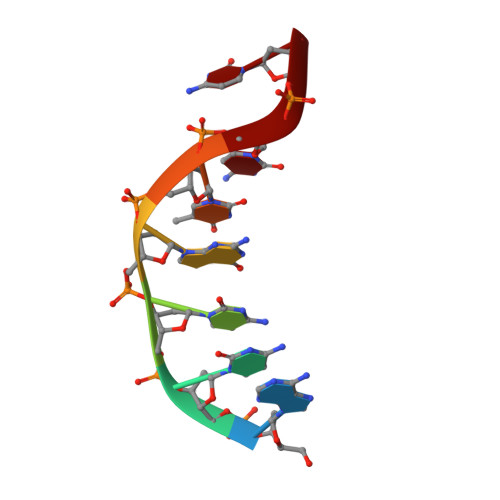
Phosphate steering by Flap Endonuclease 1 promotes 5'-flap specificity and incision to prevent genome instability.
Tsutakawa, S.E., Thompson, M.J., Arvai, A.S., Neil, A.J., Shaw, S.J., Algasaier, S.I., Kim, J.C., Finger, L.D., Jardine, E., Gotham, V.J.B., Sarker, A.H., Her, M.Z., Rashid, F., Hamdan, S.M., Mirkin, S.M., Grasby, J.A., Tainer, J.A.(2017) Nat Commun 8: 15855-15855
- PubMed: 28653660
- DOI: https://doi.org/10.1038/ncomms15855
- Primary Citation of Related Structures:
5K97, 5KSE, 5UM9 - PubMed Abstract:
DNA replication and repair enzyme Flap Endonuclease 1 (FEN1) is vital for genome integrity, and FEN1 mutations arise in multiple cancers. FEN1 precisely cleaves single-stranded (ss) 5'-flaps one nucleotide into duplex (ds) DNA. Yet, how FEN1 selects for but does not incise the ss 5'-flap was enigmatic. Here we combine crystallographic, biochemical and genetic analyses to show that two dsDNA binding sites set the 5'polarity and to reveal unexpected control of the DNA phosphodiester backbone by electrostatic interactions. Via 'phosphate steering', basic residues energetically steer an inverted ss 5'-flap through a gateway over FEN1's active site and shift dsDNA for catalysis. Mutations of these residues cause an 18,000-fold reduction in catalytic rate in vitro and large-scale trinucleotide (GAA) n repeat expansions in vivo, implying failed phosphate-steering promotes an unanticipated lagging-strand template-switch mechanism during replication. Thus, phosphate steering is an unappreciated FEN1 function that enforces 5'-flap specificity and catalysis, preventing genomic instability.
- Molecular Biophysics and Integrated Bioimaging, Lawrence Berkeley National Laboratory, Berkeley, California 94720, USA.
Organizational Affiliation: