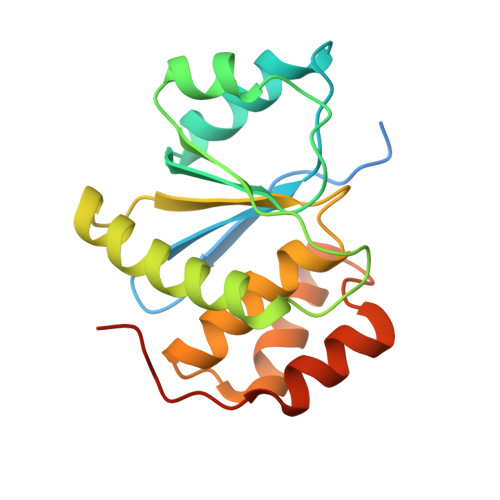
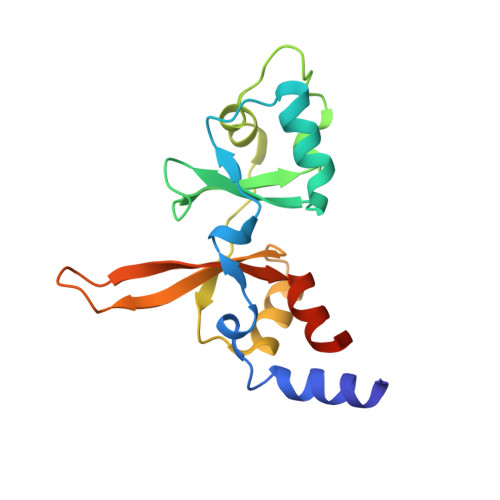
Phosphocysteine in the PRL-CNNM pathway mediates magnesium homeostasis.
Gulerez, I., Funato, Y., Wu, H., Yang, M., Kozlov, G., Miki, H., Gehring, K.(2016) EMBO Rep 17: 1890-1900
- PubMed: 27856537
- DOI: https://doi.org/10.15252/embr.201643393
- Primary Citation of Related Structures:
5K22 - PubMed Abstract:
PRLs (phosphatases of regenerating liver) are frequently overexpressed in human cancers and are prognostic markers of poor survival. Despite their potential as therapeutic targets, their mechanism of action is not understood in part due to their weak enzymatic activity. Previous studies revealed that PRLs interact with CNNM ion transporters and prevent CNNM4-dependent Mg 2+ transport, which is important for energy metabolism and tumor progression. Here, we report that PRL-CNNM complex formation is regulated by the formation of phosphocysteine. We show that cysteine in the PRL catalytic site is endogenously phosphorylated as part of the catalytic cycle and that phosphocysteine levels change in response to Mg 2+ levels. Phosphorylation blocks PRL binding to CNNM Mg 2+ transporters, and mutations that block the PRL-CNNM interaction prevent regulation of Mg 2+ efflux in cultured cells. The crystal structure of the complex of PRL2 and the CBS-pair domain of the Mg 2+ transporter CNNM3 reveals the molecular basis for the interaction. The identification of phosphocysteine as a regulatory modification opens new perspectives for signaling by protein phosphatases.
- Department of Biochemistry and Groupe de recherche axé sur la structure des protéines, McGill University, Montreal, Quebec, Canada.
Organizational Affiliation: