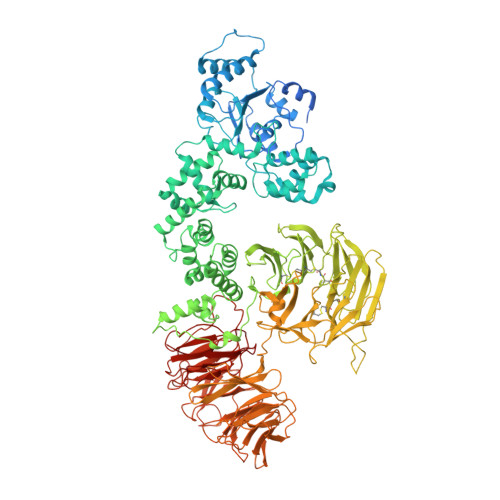
A near atomic structure of the active human apoptosome.
Cheng, T.C., Hong, C., Akey, I.V., Yuan, S., Akey, C.W.(2016) Elife 5
- PubMed: 27697150
- DOI: https://doi.org/10.7554/eLife.17755
- Primary Citation of Related Structures:
5JUY - PubMed Abstract:
In response to cell death signals, an active apoptosome is assembled from Apaf-1 and procaspase-9 (pc-9). Here we report a near atomic structure of the active human apoptosome determined by cryo-electron microscopy. The resulting model gives insights into cytochrome c binding, nucleotide exchange and conformational changes that drive assembly. During activation an acentric disk is formed on the central hub of the apoptosome. This disk contains four Apaf-1/pc-9 CARD pairs arranged in a shallow spiral with the fourth pc-9 CARD at lower occupancy. On average, Apaf-1 CARDs recruit 3 to 5 pc-9 molecules to the apoptosome and one catalytic domain may be parked on the hub, when an odd number of zymogens are bound. This suggests a stoichiometry of one or at most, two pc-9 dimers per active apoptosome. Thus, our structure provides a molecular framework to understand the role of the apoptosome in programmed cell death and disease.
- Department of Physiology and Biophysics, Boston University School of Medicine, Boston, United States.
Organizational Affiliation: