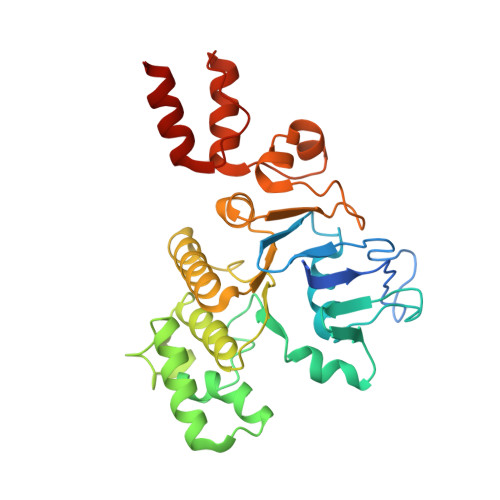
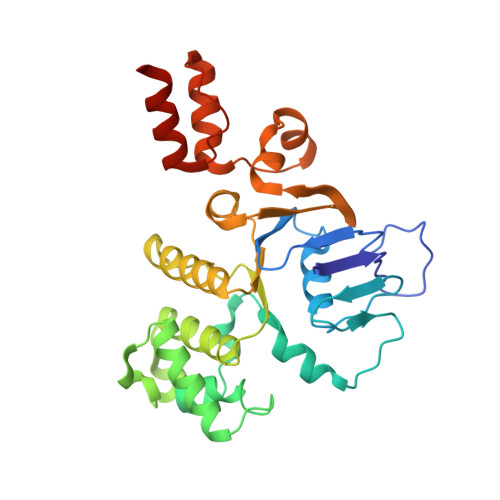
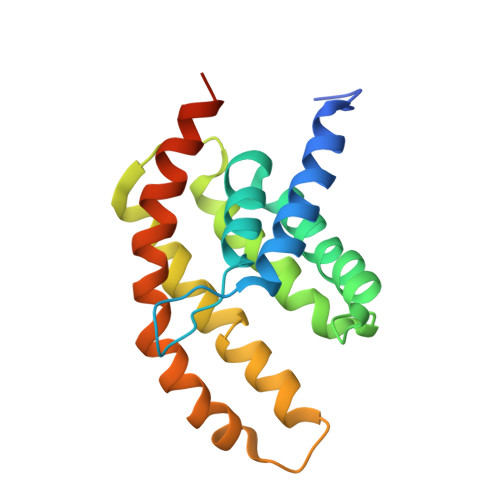
Structural insight in the toppling mechanism of an energy-coupling factor transporter.
Swier, L.J., Guskov, A., Slotboom, D.J.(2016) Nat Commun 7: 11072
- PubMed: 27026363
- DOI: https://doi.org/10.1038/ncomms11072
- Primary Citation of Related Structures:
5D0Y, 5D3M, 5JSZ - PubMed Abstract:
Energy-coupling factor (ECF) transporters mediate uptake of micronutrients in prokaryotes. The transporters consist of an S-component that binds the transported substrate and an ECF module (EcfAA'T) that binds and hydrolyses ATP. The mechanism of transport is poorly understood but presumably involves an unusual step in which the membrane-embedded S-component topples over to carry the substrate across the membrane. In many ECF transporters, the S-component dissociates from the ECF module after transport. Subsequently, substrate-bound S-components out-compete the empty proteins for re-binding to the ECF module in a new round of transport. Here we present crystal structures of the folate-specific transporter ECF-FolT from Lactobacillus delbrueckii. Interaction of the ECF module with FolT stabilizes the toppled state, and simultaneously destroys the high-affinity folate-binding site, allowing substrate release into the cytosol. We hypothesize that differences in the kinetics of toppling can explain how substrate-loaded FolT out-competes apo-FolT for association with the ECF module.
- Groningen Biomolecular Science and Biotechnology Institute, University of Groningen, Nijenborgh 4, 9747 AG Groningen, The Netherlands.
Organizational Affiliation: