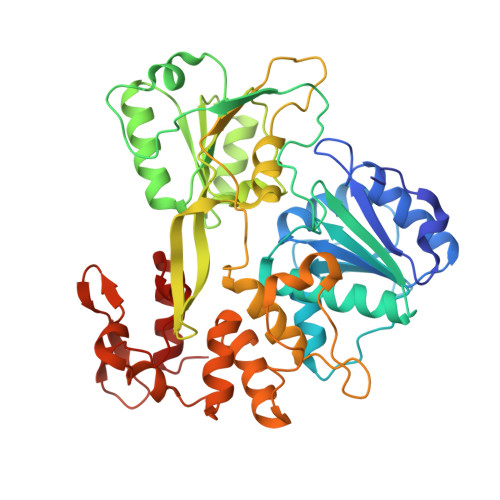
Structure of the NS3 helicase from Zika virus.
Jain, R., Coloma, J., Garcia-Sastre, A., Aggarwal, A.K.(2016) Nat Struct Mol Biol 23: 752-754
- PubMed: 27399257
- DOI: https://doi.org/10.1038/nsmb.3258
- Primary Citation of Related Structures:
5JRZ - PubMed Abstract:
Zika virus has emerged as a pathogen of major health concern. Here, we present a high-resolution (1.62-Å) crystal structure of the RNA helicase from the French Polynesia strain. The structure is similar to that of the RNA helicase from Dengue virus, with variability in the conformations of loops typically involved in binding ATP and RNA. We identify druggable 'hotspots' that are well suited for in silico and/or fragment-based high-throughput drug discovery.
- Department of Structural &Chemical Biology, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Organizational Affiliation: