Crystal structure of TgCDPK1 bound to NVPACU106
El Bakkouri, M., Walker, J.R., Loppnau, P., Graslund, S., Bountra, C., Arrowsmith, C.H., Edwards, A.M., Hui, R., Lovato, D.V., Structural Genomics Consortium (SGC)To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Calmodulin-domain protein kinase 1 | 525 | Toxoplasma gondii | Mutation(s): 0 Gene Names: CDPK1 EC: 2.7.11.1 | 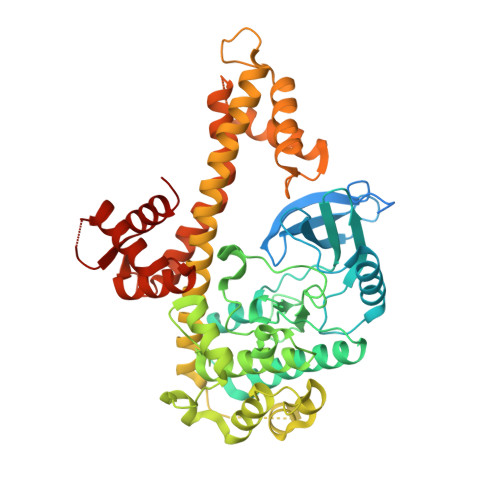 | |
UniProt | |||||
Find proteins for Q9BJF5 (Toxoplasma gondii) Explore Q9BJF5 Go to UniProtKB: Q9BJF5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9BJF5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 6LO Query on 6LO | B [auth A] | N-[(3-{4-amino-5-[3-(benzyloxy)phenyl]-7H-pyrrolo[2,3-d]pyrimidin-7-yl}cyclobutyl)methyl]acetamide C26 H27 N5 O2 BUCAFRXFNHCHRU-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | D [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| CA Query on CA | C [auth A] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 48.142 | α = 90 |
| b = 72.473 | β = 98.63 |
| c = 65.101 | γ = 90 |
| Software Name | Purpose |
|---|---|
| DENZO | data reduction |
| SCALEPACK | data scaling |
| PDB_EXTRACT | data extraction |
| BUSTER | refinement |
| HKL-3000 | data reduction |
| REFMAC | phasing |