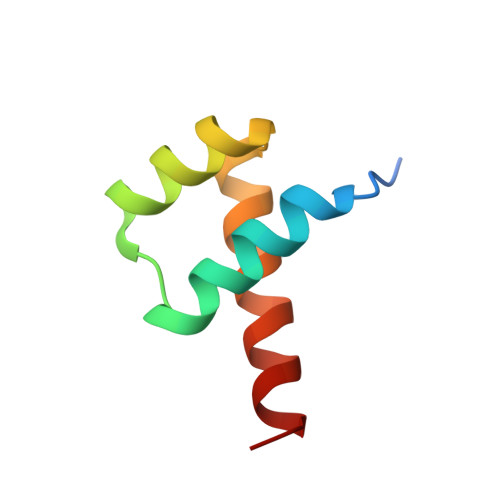
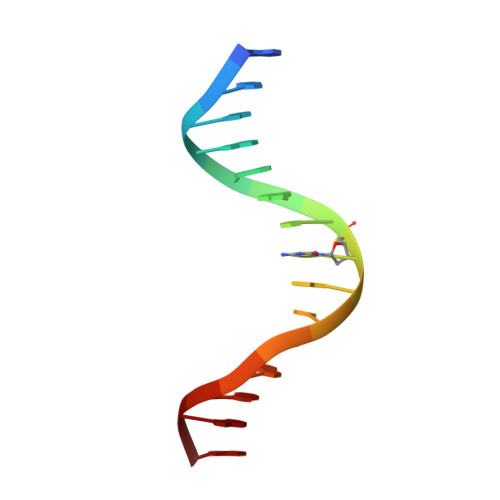
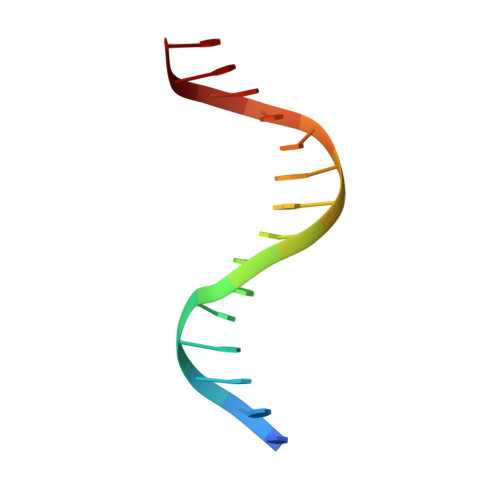
Stereospecific Effects of Oxygen-to-Sulfur Substitution in DNA Phosphate on Ion Pair Dynamics and Protein-DNA Affinity.
Nguyen, D., Zandarashvili, L., White, M.A., Iwahara, J.(2016) Chembiochem 17: 1636-1642
- PubMed: 27271797
- DOI: https://doi.org/10.1002/cbic.201600265
- Primary Citation of Related Structures:
5JLW, 5JLX - PubMed Abstract:
Oxygen-to-sulfur substitutions in DNA phosphate often enhance affinity for DNA-binding proteins. Our previous studies have suggested that this effect of sulfur substitution of both OP1 and OP2 atoms is due to an entropic gain associated with enhanced ion pair dynamics. In this work, we studied stereospecific effects of single sulfur substitution of either the OP1 or OP2 atom in DNA phosphate at the Lys57 interaction site of the Antennapedia homeodomain-DNA complex. Using crystallography, we obtained structural information on the RP and SP diastereomers of the phosphoromonothioate and their interaction with Lys57. Using fluorescence-based assays, we found significant affinity enhancement upon sulfur substitution of the OP2 atom. Using NMR spectroscopy, we found significant mobilization of the Lys57 side-chain NH3 (+) group upon sulfur substitution of the OP2 atom. These data provide further mechanistic insights into the affinity enhancement by oxygen-to-sulfur substitution in DNA phosphate.
- Department of Biochemistry and Molecular Biology, Sealy Center for Structural Biology and Molecular Biophysics, University of Texas Medical Branch, 301 University Boulevard, Medical Research Building 5.104C, Galveston, TX, 77555-1068, USA.
Organizational Affiliation: