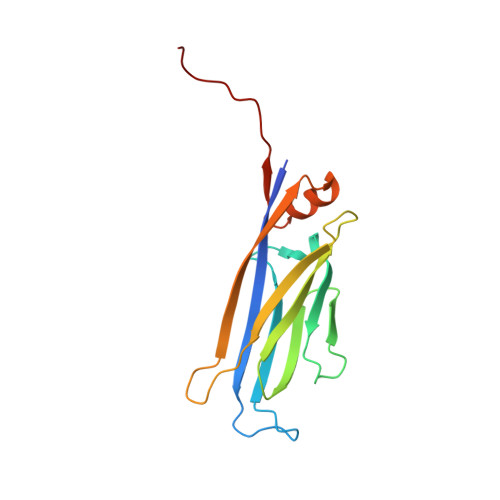
ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia
Wan, L., Wen, H., Li, Y., Lyu, J., Xi, Y., Hoshii, T., Joseph, J.K., Wang, X., Loh, Y.E., Erb, M.A., Souza, A.L., Bradner, J.E., Shen, L., Li, W., Li, H., Allis, C.D., Armstrong, S.A., Shi, X.(2017) Nature 543: 265-269
- PubMed: 28241141
- DOI: https://doi.org/10.1038/nature21687
- Primary Citation of Related Structures:
5J9S - PubMed Abstract:
Cancer cells are characterized by aberrant epigenetic landscapes and often exploit chromatin machinery to activate oncogenic gene expression programs. Recognition of modified histones by 'reader' proteins constitutes a key mechanism underlying these processes; therefore, targeting such pathways holds clinical promise, as exemplified by the development of bromodomain and extra-terminal (BET) inhibitors. We recently identified the YEATS domain as an acetyl-lysine-binding module, but its functional importance in human cancer remains unknown. Here we show that the YEATS domain-containing protein ENL, but not its paralogue AF9, is required for disease maintenance in acute myeloid leukaemia. CRISPR-Cas9-mediated depletion of ENL led to anti-leukaemic effects, including increased terminal myeloid differentiation and suppression of leukaemia growth in vitro and in vivo. Biochemical and crystal structural studies and chromatin-immunoprecipitation followed by sequencing analyses revealed that ENL binds to acetylated histone H3, and co-localizes with H3K27ac and H3K9ac on the promoters of actively transcribed genes that are essential for leukaemia. Disrupting the interaction between the YEATS domain and histone acetylation via structure-based mutagenesis reduced the recruitment of RNA polymerase II to ENL-target genes, leading to the suppression of oncogenic gene expression programs. Notably, disrupting the functionality of ENL further sensitized leukaemia cells to BET inhibitors. Together, our data identify ENL as a histone acetylation reader that regulates oncogenic transcriptional programs in acute myeloid leukaemia, and suggest that displacement of ENL from chromatin may be a promising epigenetic therapy, alone or in combination with BET inhibitors, for aggressive leukaemia.
- Laboratory of Chromatin Biology &Epigenetics, The Rockefeller University, New York, New York 10065, USA.
Organizational Affiliation: