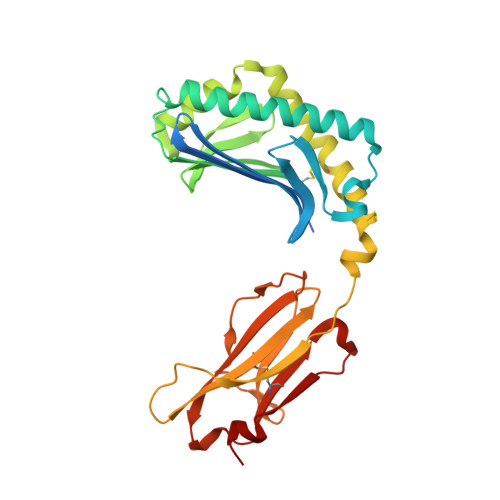
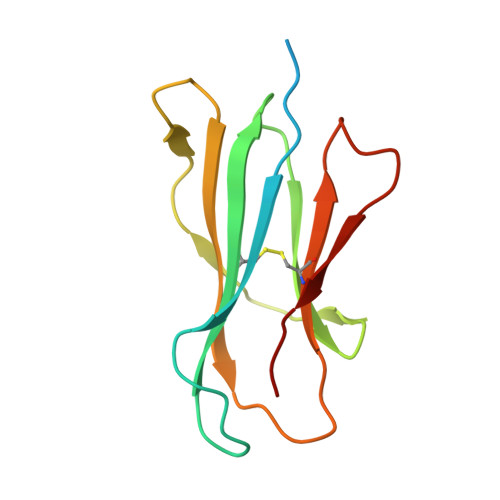
CD1a on Langerhans cells controls inflammatory skin disease.
Kim, J.H., Hu, Y., Yongqing, T., Kim, J., Hughes, V.A., Le Nours, J., Marquez, E.A., Purcell, A.W., Wan, Q., Sugita, M., Rossjohn, J., Winau, F.(2016) Nat Immunol 17: 1159-1166
- PubMed: 27548435
- DOI: https://doi.org/10.1038/ni.3523
- Primary Citation of Related Structures:
5J1A - PubMed Abstract:
CD1a is a lipid-presenting molecule that is abundantly expressed on Langerhans cells. However, the in vivo role of CD1a has remained unclear, principally because CD1a is lacking in mice. Through the use of mice with transgenic expression of CD1a, we found that the plant-derived lipid urushiol triggered CD1a-dependent skin inflammation driven by CD4(+) helper T cells that produced the cytokines IL-17 and IL-22 (TH17 cells). Human subjects with poison-ivy dermatitis had a similar cytokine signature following CD1a-mediated recognition of urushiol. Among various urushiol congeners, we identified diunsaturated pentadecylcatechol (C15:2) as the dominant antigen for CD1a-restricted T cells. We determined the crystal structure of the CD1a-urushiol (C15:2) complex, demonstrating the molecular basis of urushiol interaction with the antigen-binding cleft of CD1a. In a mouse model and in patients with psoriasis, CD1a amplified inflammatory responses that were mediated by TH17 cells that reacted to self lipid antigens. Treatment with blocking antibodies to CD1a alleviated skin inflammation. Thus, we propose CD1a as a potential therapeutic target in inflammatory skin diseases.
- Program in Cellular and Molecular Medicine, Boston Children's Hospital, Department of Microbiology and Immunobiology, Harvard Medical School, Boston, Massachusetts, USA.
Organizational Affiliation: