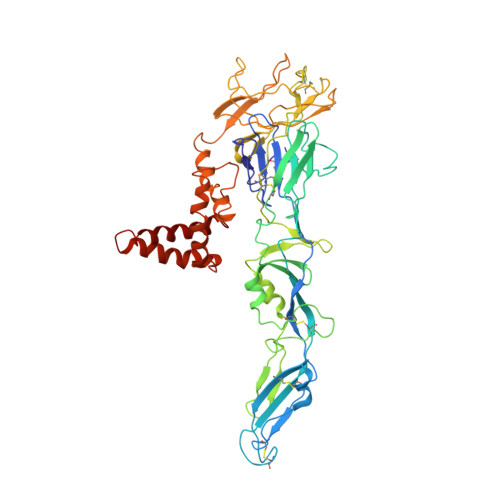
Structure of the thermally stable Zika virus
Kostyuchenko, V.A., Lim, E.X., Zhang, S., Fibriansah, G., Ng, T.S., Ooi, J.S., Shi, J., Lok, S.M.(2016) Nature 533: 425-428
- PubMed: 27093288
- DOI: https://doi.org/10.1038/nature17994
- Primary Citation of Related Structures:
5IZ7 - PubMed Abstract:
Zika virus (ZIKV), formerly a neglected pathogen, has recently been associated with microcephaly in fetuses, and with Guillian-Barré syndrome in adults. Here we present the 3.7 Å resolution cryo-electron microscopy structure of ZIKV, and show that the overall architecture of the virus is similar to that of other flaviviruses. Sequence and structural comparisons of the ZIKV envelope (E) protein with other flaviviruses show that parts of the E protein closely resemble the neurovirulent West Nile and Japanese encephalitis viruses, while others are similar to dengue virus (DENV). However, the contribution of the E protein to flavivirus pathobiology is currently not understood. The virus particle was observed to be structurally stable even when incubated at 40 °C, in sharp contrast to the less thermally stable DENV. This is also reflected in the infectivity of ZIKV compared to DENV serotypes 2 and 4 (DENV2 and DENV4) at different temperatures. The cryo-electron microscopy structure shows a virus with a more compact surface. This structural stability of the virus may help it to survive in the harsh conditions of semen, saliva and urine. Antibodies or drugs that destabilize the structure may help to reduce the disease outcome or limit the spread of the virus.
- Program in Emerging Infectious Diseases, Duke-National University of Singapore Medical School, Singapore 169857, Singapore.
Organizational Affiliation: