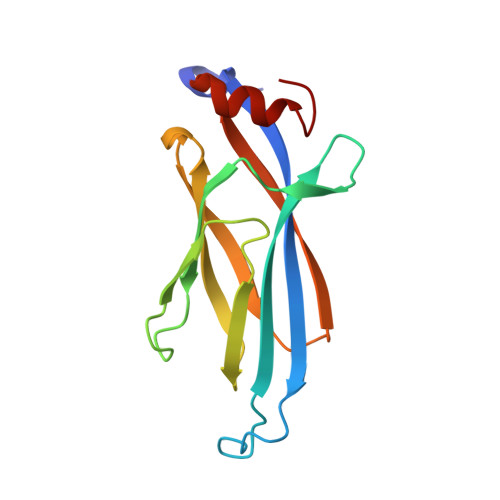
The Taf14 YEATS domain is a reader of histone crotonylation.
Andrews, F.H., Shinsky, S.A., Shanle, E.K., Bridgers, J.B., Gest, A., Tsun, I.K., Krajewski, K., Shi, X., Strahl, B.D., Kutateladze, T.G.(2016) Nat Chem Biol 12: 396-398
- PubMed: 27089029
- DOI: https://doi.org/10.1038/nchembio.2065
- Primary Citation of Related Structures:
5IOK - PubMed Abstract:
The discovery of new histone modifications is unfolding at startling rates; however, the identification of effectors capable of interpreting these modifications has lagged behind. Here we report the YEATS domain as an effective reader of histone lysine crotonylation, an epigenetic signature associated with active transcription. We show that the Taf14 YEATS domain engages crotonyllysine via a unique π-π-π-stacking mechanism and that other YEATS domains have crotonyllysine-binding activity.
- Department of Pharmacology, University of Colorado School of Medicine, Aurora, Colorado, USA.
Organizational Affiliation: