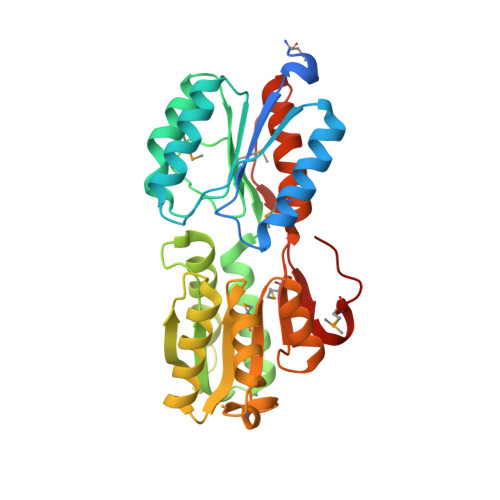
Crystal structure of ABC transporter Solute Binding Protein B1G1H7 from Burkholderia graminis C4D1M, target EFI-511179, in complex with D-arabinose
Roth, Y., Vetting, M.W., Al Obaidi, N.F., Toro, R., Morisco, L.L., Benach, J., Koss, J., Wasserman, S.R., Gerlt, J.A., Almo, S.C., Enzyme Function Initiative (EFI)To be published.