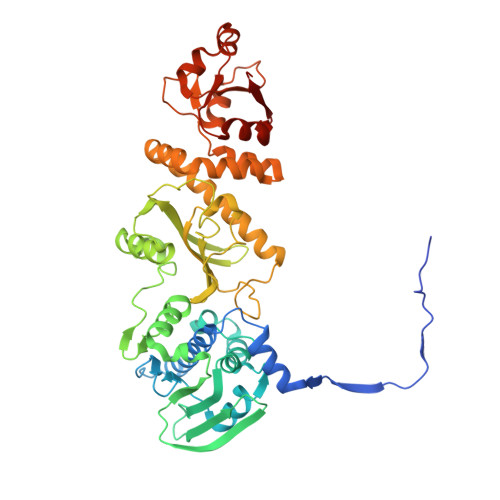
2.4 angstrom resolution crystal structure of human TRAP1NM, the Hsp90 paralog in the mitochondrial matrix.
Sung, N., Lee, J., Kim, J.H., Chang, C., Tsai, F.T., Lee, S.(2016) Acta Crystallogr D Struct Biol 72: 904-911
- PubMed: 27487821
- DOI: https://doi.org/10.1107/S2059798316009906
- Primary Citation of Related Structures:
5HPH - PubMed Abstract:
TRAP1 is an organelle-specific Hsp90 paralog that is essential for neoplastic growth. As a member of the Hsp90 family, TRAP1 is presumed to be a general chaperone facilitating the late-stage folding of Hsp90 client proteins in the mitochondrial matrix. Interestingly, TRAP1 cannot replace cytosolic Hsp90 in protein folding, and none of the known Hsp90 co-chaperones are found in mitochondria. Thus, the three-dimensional structure of TRAP1 must feature regulatory elements that are essential to the ATPase activity and chaperone function of TRAP1. Here, the crystal structure of a human TRAP1NM dimer is presented, featuring an intact N-domain and M-domain structure, bound to adenosine 5'-β,γ-imidotriphosphate (ADPNP). The crystal structure together with epitope-mapping results shows that the TRAP1 M-domain loop 1 contacts the neighboring subunit and forms a previously unobserved third dimer interface that mediates the specific interaction with mitochondrial Hsp70.
- Verna and Marrs McLean Department of Biochemistry and Molecular Biology, Baylor College of Medicine, Houston, TX 77030, USA.
Organizational Affiliation: